> Combine two datasets
There are six _join_ (or _merge_) options available in Radiant from the dplyr package developed by Hadley Wickham et.al.
The examples below are adapted from the Cheatsheet for dplyr join functions by Jenny Bryan and focus on three small datasets, `superheroes`, `publishers`, and `avengers`, to illustrate the different _join_ types and other ways to combine datasets in R and Radiant. The data are also available in csv format through the links below:
* superheroes.csv
* publishers.csv
* avengers.csv
```{r results = 'asis', echo = FALSE}
tab_small <- "class='table table-condensed table-hover' style='width:30%;'"
tab_big <- "class='table table-condensed table-hover' style='width:70%;'"
data(superheroes, package = "radiant.data", envir = environment())
knitr::kable(
superheroes, align = 'l', format = 'html',
caption = "Superheroes", table.attr = tab_big
)
```
```{r results = 'asis', echo = FALSE}
data(publishers, package = "radiant.data", envir = environment())
knitr::kable(
publishers, align = 'l', format = 'html',
caption = "Publishers", table.attr = tab_small
)
```
In the screen-shot of the _Data > Combine_ tab below we see the two datasets. The tables share the variable _publisher_ which is automatically selected for the join. Different join options are available from the `Combine type` dropdown. You can also specify a name for the combined dataset in the `Combined dataset` text input box.
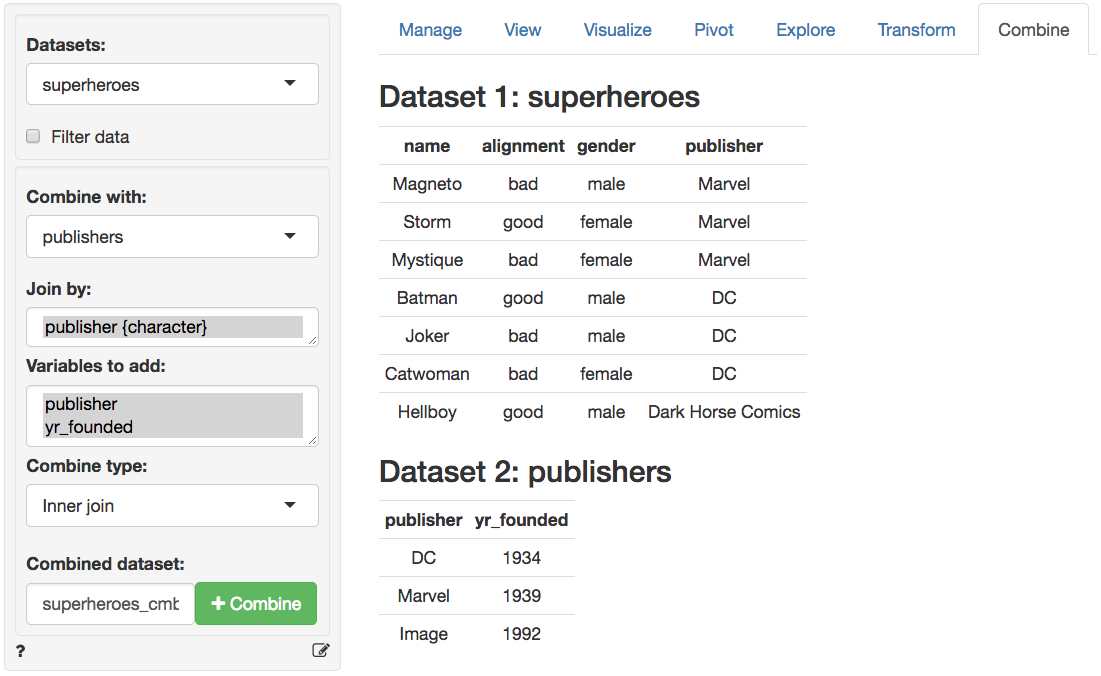
### Inner join (superheroes, publishers)
If x = superheroes and y = publishers:
> An inner join returns all rows from x with matching values in y, and all columns from both x and y. If there are multiple matches between x and y, all match combinations are returned.
```{r results = 'asis', echo = FALSE}
dplyr::inner_join(superheroes, publishers, by = "publisher") %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
In the table above we lose _Hellboy_ because, although this hero does appear in `superheroes`, the publisher (_Dark Horse Comics_) does not appear in `publishers`. The join result has all variables from `superheroes`, plus _yr\_founded_, from `publishers`. We can visualize an inner join with the venn-diagram below:
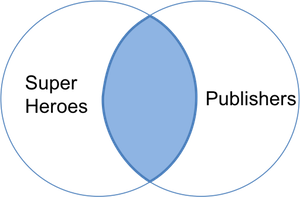
The R(adiant) commands are:
```r
# Radiant
combine_data(superheroes, publishers, by = "publisher", type = "inner_join")
# R
inner_join(superheroes, publishers, by = "publisher")
```
### Left join (superheroes, publishers)
> A left join returns all rows from x, and all columns from x and y. If there are multiple matches between x and y, all match combinations are returned.
```{r results = 'asis', echo = FALSE}
dplyr::left_join(superheroes, publishers, by = "publisher") %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
The join result contains `superheroes` with variable `yr_founded` from `publishers`. _Hellboy_, whose publisher does not appear in `publishers`, has an `NA` for _yr_founded_. We can visualize a left join with the venn-diagram below:
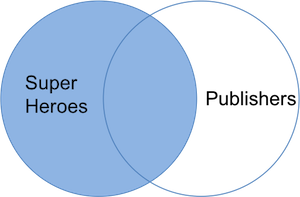
The R(adiant) commands are:
```r
# Radiant
combine_data(superheroes, publishers, by = "publisher", type = "left_join")
# R
left_join(superheroes, publishers, by = "publisher")
```
### Right join (superheroes, publishers)
> A right join returns all rows from y, and all columns from y and x. If there are multiple matches between y and x, all match combinations are returned.
```{r results = 'asis', echo = FALSE}
dplyr::right_join(superheroes, publishers, by = "publisher") %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
The join result contains all rows and columns from `publishers` and all variables from `superheroes`. We lose _Hellboy_, whose publisher does not appear in `publishers`. _Image_ is retained in the table but has `NA` values for the variables _name_, _alignment_, and _gender_ from `superheroes`. Notice that a join can change both the row and variable order so you should not rely on these in your analysis. We can visualize a right join with the venn-diagram below:
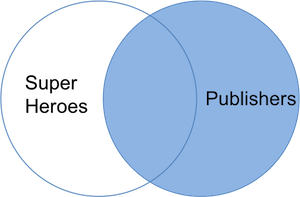
The R(adiant) commands are:
```r
# Radiant
combine_data(superheroes, publishers, by = "publisher", type = "right_join")
# R
right_join(superheroes, publishers, by = "publisher")
```
### Full join (superheroes, publishers)
> A full join combines two datasets, keeping rows and columns that appear in either.
```{r results = 'asis', echo = FALSE}
dplyr::full_join(superheroes, publishers, by = "publisher") %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
In this table we keep _Hellboy_ (even though _Dark Horse Comics_ is not in `publishers`) and _Image_ (even though the publisher is not listed in `superheroes`) and get variables from both datasets. Observations without a match are assigned the value NA for variables from the _other_ dataset. We can visualize a full join with the venn-diagram below:
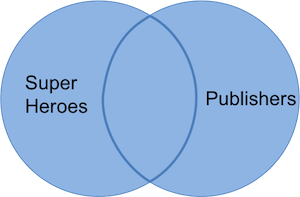
The R(adiant) commands are:
```r
# Radiant
combine_data(superheroes, publishers, by = "publisher", type = "full_join")
# R
full_join(superheroes, publishers, by = "publisher")
```
### Semi join (superheroes, publishers)
> A semi join keeps only columns from x. Whereas an inner join will return one row of x for each matching row of y, a semi join will never duplicate rows of x.
```{r results = 'asis', echo = FALSE}
dplyr::semi_join(superheroes, publishers, by = "publisher") %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
We get a similar table as with `inner_join` but it contains only the variables in `superheroes`. The R(adiant) commands are:
```r
# Radiant
combine_data(superheroes, publishers, by = "publisher", type = "semi_join")
# R
semi_join(superheroes, publishers, by = "publisher")
```
### Anti join (superheroes, publishers)
> An anti join returns all rows from x without matching values in y, keeping only columns from x
```{r results = 'asis', echo = FALSE}
dplyr::anti_join(superheroes, publishers, by = "publisher") %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
We now get **only** _Hellboy_, the only superhero not in `publishers` and we do not get the variable _yr\_founded_ either. We can visualize an anti join with the venn-diagram below:
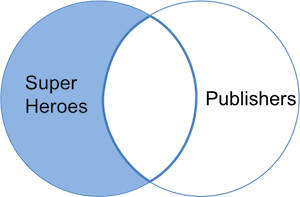
### Dataset order
Note that the order of the datasets selected may matter for a join. If we setup the _Data > Combine_ tab as below the results are as follows:
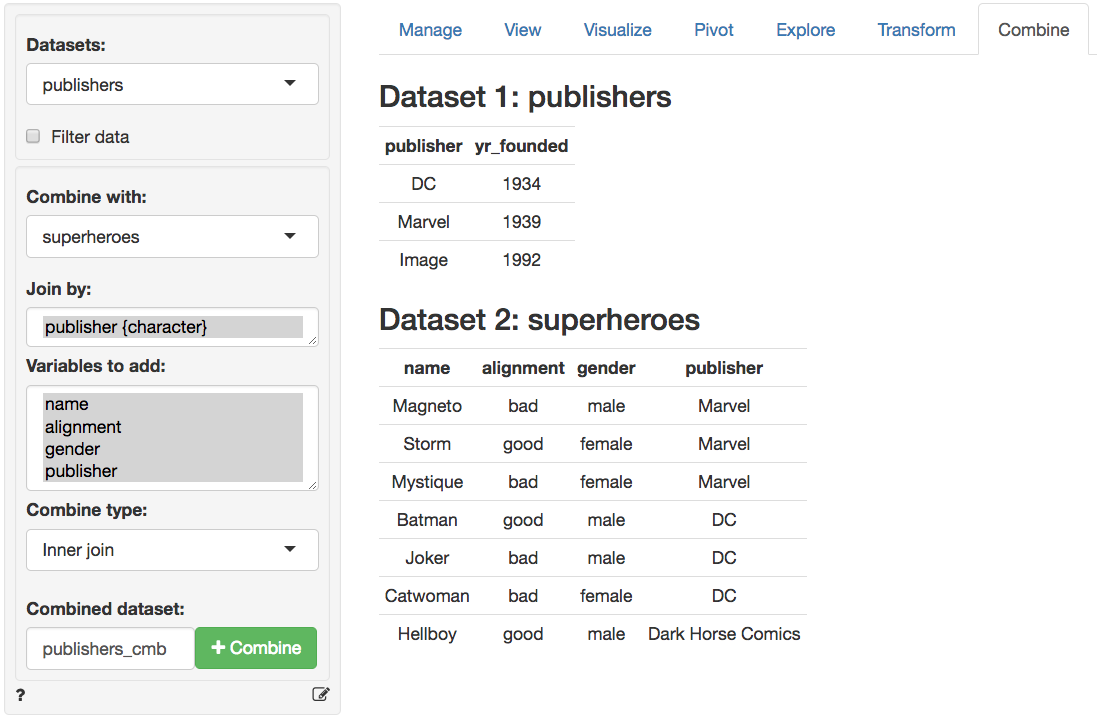
### Inner join (publishers, superheroes)
```{r results = 'asis', echo = FALSE}
dplyr::inner_join(publishers, superheroes, by = "publisher") %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
Every publisher that has a match in `superheroes` appears multiple times, once for each match. Apart from variable and row order, this is the same result we had for the inner join shown above.
### Left and Right join (publishers, superheroes)
Apart from row and variable order, a left join of `publishers` and `superheroes` is equivalent to a right join of `superheroes` and `publishers`. Similarly, a right join of `publishers` and `superheroes` is equivalent to a left join of `superheroes` and `publishers`.
### Full join (publishers, superheroes)
As you might expect, apart from row and variable order, a full join of `publishers` and `superheroes` is equivalent to a full join of `superheroes` and `publishers`.
### Semi join (publishers, superheroes)
```{r results = 'asis', echo = FALSE}
dplyr::semi_join(publishers, superheroes, by = "publisher") %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_small)
```
With semi join the effect of switching the dataset order is more clear. Even though there are multiple matches for each publisher only one is shown. Contrast this with an inner join where "If there are multiple matches between x and y, all match combinations are returned." We see that publisher _Image_ is lost in the table because it is not in `superheroes`.
### Anti join (publishers, superheroes)
```{r results = 'asis', echo = FALSE}
dplyr::anti_join(publishers, superheroes, by = "publisher") %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_small)
```
Only publisher _Image_ is retained because both _Marvel_ and _DC_ are in `superheroes`. We keep only variables in `publishers`.
### Additional tools to combine datasets (avengers, superheroes)
When two datasets have the same columns (or rows) there are additional ways in which we can combine them into a new dataset. We have already used the `superheroes` dataset and will now try to combine it with the `avengers` data. These two datasets have the same number of rows and columns and the columns have the same names.
In the screen-shot of the _Data > Combine_ tab below we see the two datasets. There is no need to select variables to combine the datasets here. Any variables in `Select variables` are ignored in the commands below. Again, you can specify a name for the combined dataset in the `Combined dataset` text input box.
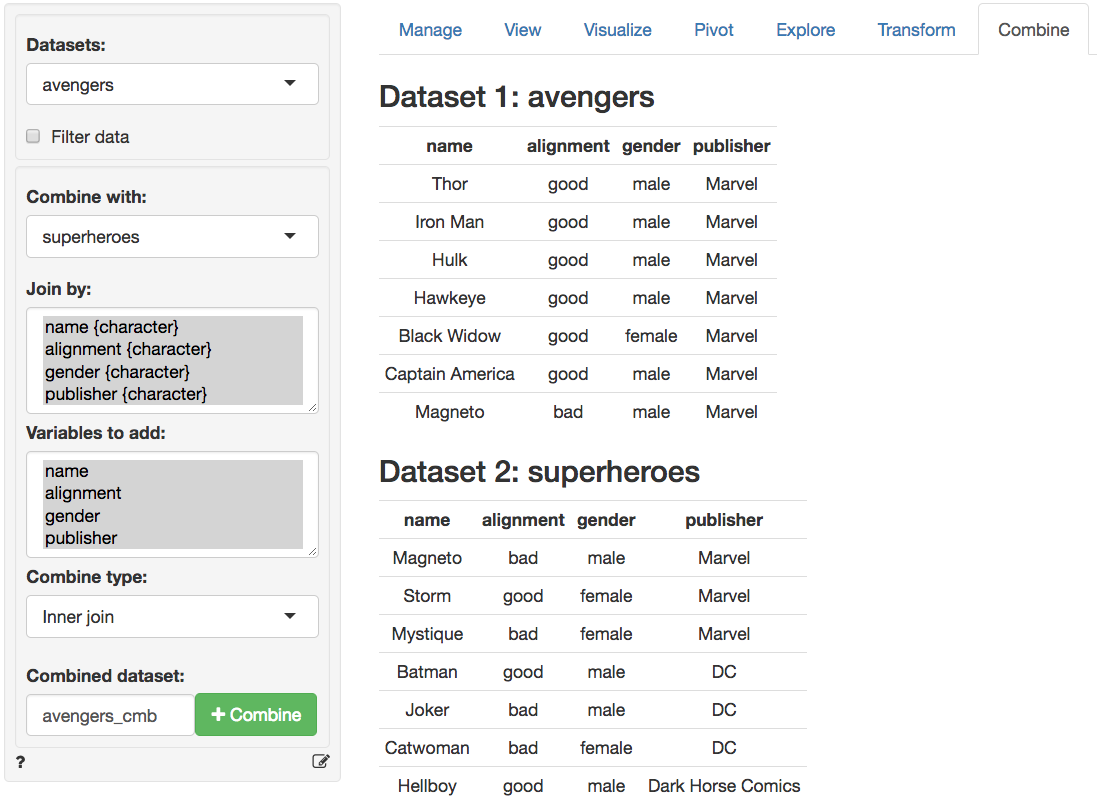
### Bind rows
```{r results = 'asis', echo = FALSE}
data(avengers, package = "radiant.data", envir = environment())
dplyr::bind_rows(avengers, superheroes) %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
If the `avengers` dataset were meant to extend the list of superheroes we could just stack the two datasets, one below the other. The new datasets has 14 rows and 4 columns. Due to a coding error in the `avengers` dataset (i.e.., _Magneto_ is *not* an _Avenger_) there is a duplicate row in the new combined dataset. Something we probably don't want.
The R(adiant) commands are:
```r
# Radiant
combine_data(avengers, superheroes, type = "bind_rows")
# R
bind_rows(avengers, superheroes)
```
### Bind columns
```{r results = 'asis', echo = FALSE}
dplyr::bind_cols(avengers, superheroes) %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
If the dataset had different columns for the same superheroes we could combine the two datasets, side by side. In radiant you will see an error message if you try to bind these columns because they have the same name. Something that we should always avoid. The method can be useful if we *know* the order of the row ids of two dataset are the same but the columns are all different.
### Intersect
```{r results = 'asis', echo = FALSE}
dplyr::intersect(avengers, superheroes) %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
A good way to check if two datasets with the same columns have duplicate rows is to choose `intersect` from the `Combine type` dropdown. There is indeed one row that is identical in the `avengers` and `superheroes` data (i.e., _Magneto_).
The R(adiant) commands are the same as shown above, except you will need to replace `bind_rows` by `intersect`.
### Union
```{r results = 'asis', echo = FALSE}
dplyr::union(avengers, superheroes) %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
A `union` of `avengers` and `superheroes` will combine the datasets but will omit duplicate rows (i.e., it will keep only one _copy_ of the row for _Magneto_). Likely what we want here.
The R(adiant) commands are the same as shown above, except you will need to replace `bind_rows` by `union`.
### Setdiff
```{r results = 'asis', echo = FALSE}
dplyr::setdiff(avengers, superheroes) %>%
knitr::kable(., align = 'l', format = 'html', table.attr = tab_big)
```
Finally, a `setdiff` will keep rows from `avengers` that are _not_ in `superheroes`. If we reverse the inputs (i.e., choose `superheroes` from the `Datasets` dropdown and `superheroes` from the `Combine with` dropdown) we will end up with all rows from `superheroes` that are not in `avengers`. In both cases the entry for _Magneto_ will be omitted.
The R(adiant) commands are the same as shown above, except you will need to replace `bind_rows` by `setdiff`.
### Report > Rmd
Add code to _Report > Rmd_ to (re)create the combined dataset by clicking the icon on the bottom left of your screen or by pressing `ALT-enter` on your keyboard.
For additional discussion see the chapter on relational data in R for data science and Tidy Explain
### R-functions
For help with the `combine_data` function see _Data > Combine_








