 \ No newline at end of file
diff --git a/radiant.basics/_pkgdown.yml b/radiant.basics/_pkgdown.yml
new file mode 100644
index 0000000000000000000000000000000000000000..36f50234d4e4d1349a8e10057989da411cc1b35f
--- /dev/null
+++ b/radiant.basics/_pkgdown.yml
@@ -0,0 +1,136 @@
+url: https://radiant-rstats.github.io/radiant.basics
+
+template:
+ params:
+ docsearch:
+ api_key: 0629d253426ce7046f92e2bc5bb11b03
+ index_name: radiant_basics
+
+navbar:
+ title: "radiant.basics"
+ left:
+ - icon: fa-home fa-lg
+ href: index.html
+ - text: "Reference"
+ href: reference/index.html
+ - text: "Articles"
+ href: articles/index.html
+ - text: "Changelog"
+ href: news/index.html
+ - text: "Other Packages"
+ menu:
+ - text: "radiant"
+ href: https://radiant-rstats.github.io/radiant/
+ - text: "radiant.data"
+ href: https://radiant-rstats.github.io/radiant.data/
+ - text: "radiant.design"
+ href: https://radiant-rstats.github.io/radiant.design/
+ - text: "radiant.basics"
+ href: https://radiant-rstats.github.io/radiant.basics/
+ - text: "radiant.model"
+ href: https://radiant-rstats.github.io/radiant.model/
+ - text: "radiant.multivariate"
+ href: https://radiant-rstats.github.io/radiant.multivariate/
+ - text: "docker"
+ href: https://github.com/radiant-rstats/docker
+ right:
+ - icon: fa-twitter fa-lg
+ href: https://twitter.com/vrnijs
+ - icon: fa-github fa-lg
+ href: https://github.com/radiant-rstats
+
+reference:
+- title: Basics > Probability
+ desc: Functions used with the Probability Calculator and the Central Limit Theorem simulator
+ contents:
+ - clt
+ - plot.clt
+ - prob_binom
+ - summary.prob_binom
+ - plot.prob_binom
+ - prob_chisq
+ - summary.prob_chisq
+ - plot.prob_chisq
+ - prob_disc
+ - summary.prob_disc
+ - plot.prob_disc
+ - prob_expo
+ - summary.prob_expo
+ - plot.prob_expo
+ - prob_fdist
+ - summary.prob_fdist
+ - plot.prob_fdist
+ - prob_lnorm
+ - summary.prob_lnorm
+ - plot.prob_lnorm
+ - prob_norm
+ - summary.prob_norm
+ - plot.prob_norm
+ - prob_pois
+ - summary.prob_pois
+ - plot.prob_pois
+ - prob_tdist
+ - summary.prob_tdist
+ - plot.prob_tdist
+ - prob_unif
+ - summary.prob_unif
+ - plot.prob_unif
+- title: Basics > Means
+ desc: Functions used with Basics > Means
+ contents:
+ - single_mean
+ - summary.single_mean
+ - plot.single_mean
+ - compare_means
+ - summary.compare_means
+ - plot.compare_means
+- title: Basics > Proportions
+ desc: Functions used with Basics > Proportions
+ contents:
+ - single_prop
+ - summary.single_prop
+ - plot.single_prop
+ - compare_props
+ - summary.compare_props
+ - plot.compare_props
+- title: Basics > Tables
+ desc: Functions used with Basics > Tables
+ contents:
+ - goodness
+ - summary.goodness
+ - plot.goodness
+ - cross_tabs
+ - summary.cross_tabs
+ - plot.cross_tabs
+ - correlation
+ - summary.correlation
+ - plot.correlation
+ - print.rcorr
+ - cor2df
+- title: Data sets
+ desc: Data sets bundled with radiant.basics
+ contents:
+ - consider
+ - demand_uk
+ - newspaper
+ - salary
+- title: Starting radiant.basics
+ desc: Functions used to start the radiant.basics shiny app
+ contents:
+ - radiant.basics
+ - radiant.basics_viewer
+ - radiant.basics_window
+articles:
+- title: Basics Menu
+ desc: >
+ These vignettes provide an introduction to the Basics menu in radiant
+ contents:
+ - pkgdown/clt
+ - pkgdown/prob_calc
+ - pkgdown/single_mean
+ - pkgdown/compare_means
+ - pkgdown/single_prop
+ - pkgdown/compare_props
+ - pkgdown/goodness
+ - pkgdown/cross_tabs
+ - pkgdown/correlation
diff --git a/radiant.basics/build/build.R b/radiant.basics/build/build.R
new file mode 100644
index 0000000000000000000000000000000000000000..c901d8c63ae7fea5f8f38df2565e82c615cc4e64
--- /dev/null
+++ b/radiant.basics/build/build.R
@@ -0,0 +1,87 @@
+setwd(rstudioapi::getActiveProject())
+curr <- getwd()
+pkg <- basename(curr)
+
+## building package for mac and windows
+rv <- R.Version()
+rv <- paste(rv$major, substr(rv$minor, 1, 1), sep = ".")
+
+rvprompt <- readline(prompt = paste0("Running for R version: ", rv, ". Is that what you wanted y/n: "))
+if (grepl("[nN]", rvprompt)) stop("Change R-version")
+
+dirsrc <- "../minicran/src/contrib"
+
+if (rv < "3.4") {
+ dirmac <- fs::path("../minicran/bin/macosx/mavericks/contrib", rv)
+} else if (rv > "3.6") {
+ dirmac <- c(
+ fs::path("../minicran/bin/macosx/big-sur-arm64/contrib", rv),
+ fs::path("../minicran/bin/macosx/contrib", rv)
+ )
+} else {
+ dirmac <- fs::path("../minicran/bin/macosx/el-capitan/contrib", rv)
+}
+
+dirwin <- fs::path("../minicran/bin/windows/contrib", rv)
+
+if (!fs::file_exists(dirsrc)) fs::dir_create(dirsrc, recursive = TRUE)
+for (d in dirmac) {
+ if (!fs::file_exists(d)) fs::dir_create(d, recursive = TRUE)
+}
+if (!fs::file_exists(dirwin)) fs::dir_create(dirwin, recursive = TRUE)
+
+# delete older version of radiant
+rem_old <- function(pkg) {
+ unlink(paste0(dirsrc, "/", pkg, "*"))
+ for (d in dirmac) {
+ unlink(paste0(d, "/", pkg, "*"))
+ }
+ unlink(paste0(dirwin, "/", pkg, "*"))
+}
+
+sapply(pkg, rem_old)
+
+## avoid 'loaded namespace' stuff when building for mac
+system(paste0(Sys.which("R"), " -e \"setwd('", getwd(), "'); app <- '", pkg, "'; source('build/build_mac.R')\""))
+
+
+win <- readline(prompt = "Did you build on Windows? y/n: ")
+if (grepl("[yY]", win)) {
+
+ fl <- list.files(pattern = "*.zip", path = "~/Dropbox/r-packages/", full.names = TRUE)
+ for (f in fl) {
+ file.copy(f, "~/gh/")
+ }
+
+ ## move packages to radiant_miniCRAN. must package in Windows first
+ # path <- normalizePath("../")
+ pth <- fs::path_abs("../")
+
+ sapply(list.files(pth, pattern = "*.tar.gz", full.names = TRUE), file.copy, dirsrc)
+ unlink("../*.tar.gz")
+ for (d in dirmac) {
+ sapply(list.files(pth, pattern = "*.tgz", full.names = TRUE), file.copy, d)
+ }
+ unlink("../*.tgz")
+ sapply(list.files(pth, pattern = "*.zip", full.names = TRUE), file.copy, dirwin)
+ unlink("../*.zip")
+
+ tools::write_PACKAGES(dirwin, type = "win.binary")
+ for (d in dirmac) {
+ tools::write_PACKAGES(d, type = "mac.binary")
+ }
+ tools::write_PACKAGES(dirsrc, type = "source")
+
+ # commit to repo
+ setwd("../minicran")
+ system("git add --all .")
+ mess <- paste0(pkg, " package update: ", format(Sys.Date(), format = "%m-%d-%Y"))
+ system(paste0("git commit -m '", mess, "'"))
+ system("git push")
+}
+
+setwd(curr)
+
+# remove.packages(c("radiant.model", "radiant.data"))
+# radiant.update::radiant.update()
+# install.packages("radiant.update")
diff --git a/radiant.basics/build/build_mac.R b/radiant.basics/build/build_mac.R
new file mode 100644
index 0000000000000000000000000000000000000000..1452bac080e154c24c6cd9acb6eef6c09a76c6ae
--- /dev/null
+++ b/radiant.basics/build/build_mac.R
@@ -0,0 +1,6 @@
+## build for mac
+app <- basename(getwd())
+curr <- setwd("../")
+f <- devtools::build(app)
+system(paste0("R CMD INSTALL --build ", f))
+setwd(curr)
diff --git a/radiant.basics/build/build_win.R b/radiant.basics/build/build_win.R
new file mode 100644
index 0000000000000000000000000000000000000000..e6861ceb5e94157a4ed21359a4d3339b9f1de8fb
--- /dev/null
+++ b/radiant.basics/build/build_win.R
@@ -0,0 +1,26 @@
+## build for windows
+rv <- R.Version()
+rv <- paste(rv$major, substr(rv$minor, 1, 1), sep = ".")
+
+rvprompt <- readline(prompt = paste0("Running for R version: ", rv, ". Is that what you wanted y/n: "))
+if (grepl("[nN]", rvprompt))
+ stop("Change R-version using Rstudio > Tools > Global Options > Rversion")
+
+## build for windows
+setwd(rstudioapi::getActiveProject())
+f <- devtools::build(binary = TRUE)
+devtools::install(upgrade = "never")
+
+fl <- list.files(pattern = "*.zip", path = "../", full.names = TRUE)
+
+for (f in fl) {
+ print(glue::glue("Copying: {f}"))
+ file.copy(f, "C:/Users/vnijs/Dropbox/r-packages/", overwrite = TRUE)
+ unlink(f)
+}
+
+#options(repos = c(RSM = "https://radiant-rstats.github.io/minicran"))
+#install.packages("radiant.data", type = "binary")
+# remove.packages(c("radiant.data", "radiant.model"))
+#install.packages("radiant.update")
+# radiant.update::radiant.update()
diff --git a/radiant.basics/data/consider.rda b/radiant.basics/data/consider.rda
new file mode 100644
index 0000000000000000000000000000000000000000..d00b31838f8bf9cbe39ca333d1fba82a47346ba9
Binary files /dev/null and b/radiant.basics/data/consider.rda differ
diff --git a/radiant.basics/data/demand_uk.rda b/radiant.basics/data/demand_uk.rda
new file mode 100644
index 0000000000000000000000000000000000000000..558d54557306b612a4faca6a794635d8e3ad3ce1
Binary files /dev/null and b/radiant.basics/data/demand_uk.rda differ
diff --git a/radiant.basics/data/newspaper.rda b/radiant.basics/data/newspaper.rda
new file mode 100644
index 0000000000000000000000000000000000000000..15cd23c5daf54ee27f71ac48adf1c6766017f220
Binary files /dev/null and b/radiant.basics/data/newspaper.rda differ
diff --git a/radiant.basics/data/salary.rda b/radiant.basics/data/salary.rda
new file mode 100644
index 0000000000000000000000000000000000000000..811a3223f58c686fb16683300103e95d6bad34b6
Binary files /dev/null and b/radiant.basics/data/salary.rda differ
diff --git a/radiant.basics/inst/app/global.R b/radiant.basics/inst/app/global.R
new file mode 100644
index 0000000000000000000000000000000000000000..f83f097571a1d7f92255ec4dcb32d6d5ebc00c5d
--- /dev/null
+++ b/radiant.basics/inst/app/global.R
@@ -0,0 +1,32 @@
+library(shiny.i18n)
+# file with translations
+i18n <- Translator$new(translation_csvs_path = "../translations")
+
+# change this to zh
+i18n$set_translation_language("zh")
+
+## sourcing from radiant.data
+options(radiant.path.data = system.file(package = "radiant.data"))
+source(file.path(getOption("radiant.path.data"), "app/global.R"), encoding = getOption("radiant.encoding", default = "UTF-8"), local = TRUE)
+
+ifelse(grepl("radiant.basics", getwd()) && file.exists("../../inst"), "..", system.file(package = "radiant.basics")) %>%
+ options(radiant.path.basics = .)
+
+## setting path for figures in help files
+addResourcePath("figures_basics", "tools/help/figures/")
+
+## setting path for www resources
+addResourcePath("www_basics", file.path(getOption("radiant.path.basics"), "app/www/"))
+
+## loading urls and ui
+source("init.R", encoding = getOption("radiant.encoding", "UTF-8"), local = TRUE)
+options(radiant.url.patterns = make_url_patterns())
+
+## if radiant.data is not in search main function from dplyr etc. won't be available
+if (!"package:radiant.basics" %in% search() &&
+ isTRUE(getOption("radiant.development")) &&
+ getOption("radiant.path.basics") == "..") {
+ options(radiant.from.package = FALSE)
+} else {
+ options(radiant.from.package = TRUE)
+}
diff --git a/radiant.basics/inst/app/help.R b/radiant.basics/inst/app/help.R
new file mode 100644
index 0000000000000000000000000000000000000000..040ebd73f3781357465085386964580480ed6beb
--- /dev/null
+++ b/radiant.basics/inst/app/help.R
@@ -0,0 +1,27 @@
+help_basics <- c(
+ "Probability calculator" = "prob_calc.md", "Central limit theorem" = "clt.md",
+ "Single mean" = "single_mean.md", "Compare means" = "compare_means.md",
+ "Single proportion" = "single_prop.md", "Compare proportions" = "compare_props.md",
+ "Goodness of fit" = "goodness.md", "Cross-tabs" = "cross_tabs.md",
+ "Correlation" = "correlation.md"
+)
+
+output$help_basics <- reactive(append_help("help_basics", file.path(getOption("radiant.path.basics"), "app/tools/help"), Rmd = TRUE))
+
+observeEvent(input$help_basics_all, {
+ help_switch(input$help_basics_all, "help_basics")
+})
+observeEvent(input$help_basics_none, {
+ help_switch(input$help_basics_none, "help_basics", help_on = FALSE)
+})
+
+help_basics_panel <- tagList(
+ wellPanel(
+ HTML(""),
+ checkboxGroupInput(
+ "help_basics", NULL, help_basics,
+ selected = state_group("help_basics"), inline = TRUE
+ )
+ )
+)
diff --git a/radiant.basics/inst/app/init.R b/radiant.basics/inst/app/init.R
new file mode 100644
index 0000000000000000000000000000000000000000..5f9513350f94f8488166992b6fbf5e26204ab920
--- /dev/null
+++ b/radiant.basics/inst/app/init.R
@@ -0,0 +1,49 @@
+## urls for menu
+r_url_list <- getOption("radiant.url.list")
+r_url_list[["Single mean"]] <-
+ list("tabs_single_mean" = list("Summary" = "basics/single-mean/", "Plot" = "basics/single-mean/plot/"))
+r_url_list[["Compare means"]] <-
+ list("tabs_compare_means" = list("Summary" = "basics/compare-means/", "Plot" = "basics/compare-means/plot/"))
+r_url_list[["Single proportion"]] <-
+ list("tabs_single_prop" = list("Summary" = "basics/single-prop/", "Plot" = "basics/single-prop/plot/"))
+r_url_list[["Compare proportions"]] <-
+ list("tabs_compare_props" = list("Summary" = "basics/compare-props/", "Plot" = "basics/compare-props/plot/"))
+r_url_list[["Goodness of fit"]] <-
+ list("tabs_goodness" = list("Summary" = "basics/goodness/", "Plot" = "basics/goodness/plot/"))
+r_url_list[["Cross-tabs"]] <-
+ list("tabs_cross_tabs" = list("Summary" = "basics/cross-tabs/", "Plot" = "basics/cross-tabs/plot/"))
+r_url_list[["Correlation"]] <-
+ list("tabs_correlation" = list("Summary" = "basics/correlation/", "Plot" = "basics`/correlation/plot/"))
+options(radiant.url.list = r_url_list)
+rm(r_url_list)
+
+## try http://127.0.0.1:3174/?url=basics/goodness/plot/&SSUID=local
+## http://127.0.0.1:7407/?url=basics/compare-means/plot/&SSUID=local-a82049
+
+## design menu
+options(
+ radiant.basics_ui =
+ tagList(
+ navbarMenu(
+ i18n$t("Basics"),
+ tags$head(
+ tags$script(src = "www_basics/js/run_return.js")
+ ),
+ i18n$t("Probability"),
+ tabPanel(i18n$t("Probability calculator"), uiOutput("prob_calc")),
+ tabPanel(i18n$t("Central Limit Theorem"), uiOutput("clt")),
+ "----", i18n$t("Means"),
+ tabPanel(i18n$t("Single mean"), uiOutput("single_mean")),
+ tabPanel(i18n$t("Compare means"), uiOutput("compare_means")),
+ tabPanel(i18n$t("Normality test"),uiOutput("normality_test")),
+ tabPanel(i18n$t("Homogeneity of variance test"),uiOutput("homo_variance_test")),
+ "----", i18n$t("Proportions"),
+ tabPanel(i18n$t("Single proportion"), uiOutput("single_prop")),
+ tabPanel(i18n$t("Compare proportions"), uiOutput("compare_props")),
+ "----", i18n$t("Tables"),
+ tabPanel(i18n$t("Goodness of fit"), uiOutput("goodness")),
+ tabPanel(i18n$t("Cross-tabs"), uiOutput("cross_tabs")),
+ tabPanel(i18n$t("Correlation"), uiOutput("correlation"))
+ )
+ )
+)
diff --git a/radiant.basics/inst/app/server.R b/radiant.basics/inst/app/server.R
new file mode 100644
index 0000000000000000000000000000000000000000..dd26d78c10b5e97367b84154896f2473ce022a9a
--- /dev/null
+++ b/radiant.basics/inst/app/server.R
@@ -0,0 +1,59 @@
+if (isTRUE(getOption("radiant.from.package"))) {
+ library(radiant.basics)
+}
+
+shinyServer(function(input, output, session) {
+
+ ## source shared functions
+ source(file.path(getOption("radiant.path.data"), "app/init.R"), encoding = getOption("radiant.encoding"), local = TRUE)
+ source(file.path(getOption("radiant.path.data"), "app/radiant.R"), encoding = getOption("radiant.encoding"), local = TRUE)
+ source("help.R", encoding = getOption("radiant.encoding"), local = TRUE)
+
+ ## help ui
+ output$help_basics_ui <- renderUI({
+ sidebarLayout(
+ sidebarPanel(
+ help_data_panel,
+ help_basics_panel,
+ uiOutput("help_text"),
+ width = 3
+ ),
+ mainPanel(
+ HTML(paste0("
\ No newline at end of file
diff --git a/radiant.basics/_pkgdown.yml b/radiant.basics/_pkgdown.yml
new file mode 100644
index 0000000000000000000000000000000000000000..36f50234d4e4d1349a8e10057989da411cc1b35f
--- /dev/null
+++ b/radiant.basics/_pkgdown.yml
@@ -0,0 +1,136 @@
+url: https://radiant-rstats.github.io/radiant.basics
+
+template:
+ params:
+ docsearch:
+ api_key: 0629d253426ce7046f92e2bc5bb11b03
+ index_name: radiant_basics
+
+navbar:
+ title: "radiant.basics"
+ left:
+ - icon: fa-home fa-lg
+ href: index.html
+ - text: "Reference"
+ href: reference/index.html
+ - text: "Articles"
+ href: articles/index.html
+ - text: "Changelog"
+ href: news/index.html
+ - text: "Other Packages"
+ menu:
+ - text: "radiant"
+ href: https://radiant-rstats.github.io/radiant/
+ - text: "radiant.data"
+ href: https://radiant-rstats.github.io/radiant.data/
+ - text: "radiant.design"
+ href: https://radiant-rstats.github.io/radiant.design/
+ - text: "radiant.basics"
+ href: https://radiant-rstats.github.io/radiant.basics/
+ - text: "radiant.model"
+ href: https://radiant-rstats.github.io/radiant.model/
+ - text: "radiant.multivariate"
+ href: https://radiant-rstats.github.io/radiant.multivariate/
+ - text: "docker"
+ href: https://github.com/radiant-rstats/docker
+ right:
+ - icon: fa-twitter fa-lg
+ href: https://twitter.com/vrnijs
+ - icon: fa-github fa-lg
+ href: https://github.com/radiant-rstats
+
+reference:
+- title: Basics > Probability
+ desc: Functions used with the Probability Calculator and the Central Limit Theorem simulator
+ contents:
+ - clt
+ - plot.clt
+ - prob_binom
+ - summary.prob_binom
+ - plot.prob_binom
+ - prob_chisq
+ - summary.prob_chisq
+ - plot.prob_chisq
+ - prob_disc
+ - summary.prob_disc
+ - plot.prob_disc
+ - prob_expo
+ - summary.prob_expo
+ - plot.prob_expo
+ - prob_fdist
+ - summary.prob_fdist
+ - plot.prob_fdist
+ - prob_lnorm
+ - summary.prob_lnorm
+ - plot.prob_lnorm
+ - prob_norm
+ - summary.prob_norm
+ - plot.prob_norm
+ - prob_pois
+ - summary.prob_pois
+ - plot.prob_pois
+ - prob_tdist
+ - summary.prob_tdist
+ - plot.prob_tdist
+ - prob_unif
+ - summary.prob_unif
+ - plot.prob_unif
+- title: Basics > Means
+ desc: Functions used with Basics > Means
+ contents:
+ - single_mean
+ - summary.single_mean
+ - plot.single_mean
+ - compare_means
+ - summary.compare_means
+ - plot.compare_means
+- title: Basics > Proportions
+ desc: Functions used with Basics > Proportions
+ contents:
+ - single_prop
+ - summary.single_prop
+ - plot.single_prop
+ - compare_props
+ - summary.compare_props
+ - plot.compare_props
+- title: Basics > Tables
+ desc: Functions used with Basics > Tables
+ contents:
+ - goodness
+ - summary.goodness
+ - plot.goodness
+ - cross_tabs
+ - summary.cross_tabs
+ - plot.cross_tabs
+ - correlation
+ - summary.correlation
+ - plot.correlation
+ - print.rcorr
+ - cor2df
+- title: Data sets
+ desc: Data sets bundled with radiant.basics
+ contents:
+ - consider
+ - demand_uk
+ - newspaper
+ - salary
+- title: Starting radiant.basics
+ desc: Functions used to start the radiant.basics shiny app
+ contents:
+ - radiant.basics
+ - radiant.basics_viewer
+ - radiant.basics_window
+articles:
+- title: Basics Menu
+ desc: >
+ These vignettes provide an introduction to the Basics menu in radiant
+ contents:
+ - pkgdown/clt
+ - pkgdown/prob_calc
+ - pkgdown/single_mean
+ - pkgdown/compare_means
+ - pkgdown/single_prop
+ - pkgdown/compare_props
+ - pkgdown/goodness
+ - pkgdown/cross_tabs
+ - pkgdown/correlation
diff --git a/radiant.basics/build/build.R b/radiant.basics/build/build.R
new file mode 100644
index 0000000000000000000000000000000000000000..c901d8c63ae7fea5f8f38df2565e82c615cc4e64
--- /dev/null
+++ b/radiant.basics/build/build.R
@@ -0,0 +1,87 @@
+setwd(rstudioapi::getActiveProject())
+curr <- getwd()
+pkg <- basename(curr)
+
+## building package for mac and windows
+rv <- R.Version()
+rv <- paste(rv$major, substr(rv$minor, 1, 1), sep = ".")
+
+rvprompt <- readline(prompt = paste0("Running for R version: ", rv, ". Is that what you wanted y/n: "))
+if (grepl("[nN]", rvprompt)) stop("Change R-version")
+
+dirsrc <- "../minicran/src/contrib"
+
+if (rv < "3.4") {
+ dirmac <- fs::path("../minicran/bin/macosx/mavericks/contrib", rv)
+} else if (rv > "3.6") {
+ dirmac <- c(
+ fs::path("../minicran/bin/macosx/big-sur-arm64/contrib", rv),
+ fs::path("../minicran/bin/macosx/contrib", rv)
+ )
+} else {
+ dirmac <- fs::path("../minicran/bin/macosx/el-capitan/contrib", rv)
+}
+
+dirwin <- fs::path("../minicran/bin/windows/contrib", rv)
+
+if (!fs::file_exists(dirsrc)) fs::dir_create(dirsrc, recursive = TRUE)
+for (d in dirmac) {
+ if (!fs::file_exists(d)) fs::dir_create(d, recursive = TRUE)
+}
+if (!fs::file_exists(dirwin)) fs::dir_create(dirwin, recursive = TRUE)
+
+# delete older version of radiant
+rem_old <- function(pkg) {
+ unlink(paste0(dirsrc, "/", pkg, "*"))
+ for (d in dirmac) {
+ unlink(paste0(d, "/", pkg, "*"))
+ }
+ unlink(paste0(dirwin, "/", pkg, "*"))
+}
+
+sapply(pkg, rem_old)
+
+## avoid 'loaded namespace' stuff when building for mac
+system(paste0(Sys.which("R"), " -e \"setwd('", getwd(), "'); app <- '", pkg, "'; source('build/build_mac.R')\""))
+
+
+win <- readline(prompt = "Did you build on Windows? y/n: ")
+if (grepl("[yY]", win)) {
+
+ fl <- list.files(pattern = "*.zip", path = "~/Dropbox/r-packages/", full.names = TRUE)
+ for (f in fl) {
+ file.copy(f, "~/gh/")
+ }
+
+ ## move packages to radiant_miniCRAN. must package in Windows first
+ # path <- normalizePath("../")
+ pth <- fs::path_abs("../")
+
+ sapply(list.files(pth, pattern = "*.tar.gz", full.names = TRUE), file.copy, dirsrc)
+ unlink("../*.tar.gz")
+ for (d in dirmac) {
+ sapply(list.files(pth, pattern = "*.tgz", full.names = TRUE), file.copy, d)
+ }
+ unlink("../*.tgz")
+ sapply(list.files(pth, pattern = "*.zip", full.names = TRUE), file.copy, dirwin)
+ unlink("../*.zip")
+
+ tools::write_PACKAGES(dirwin, type = "win.binary")
+ for (d in dirmac) {
+ tools::write_PACKAGES(d, type = "mac.binary")
+ }
+ tools::write_PACKAGES(dirsrc, type = "source")
+
+ # commit to repo
+ setwd("../minicran")
+ system("git add --all .")
+ mess <- paste0(pkg, " package update: ", format(Sys.Date(), format = "%m-%d-%Y"))
+ system(paste0("git commit -m '", mess, "'"))
+ system("git push")
+}
+
+setwd(curr)
+
+# remove.packages(c("radiant.model", "radiant.data"))
+# radiant.update::radiant.update()
+# install.packages("radiant.update")
diff --git a/radiant.basics/build/build_mac.R b/radiant.basics/build/build_mac.R
new file mode 100644
index 0000000000000000000000000000000000000000..1452bac080e154c24c6cd9acb6eef6c09a76c6ae
--- /dev/null
+++ b/radiant.basics/build/build_mac.R
@@ -0,0 +1,6 @@
+## build for mac
+app <- basename(getwd())
+curr <- setwd("../")
+f <- devtools::build(app)
+system(paste0("R CMD INSTALL --build ", f))
+setwd(curr)
diff --git a/radiant.basics/build/build_win.R b/radiant.basics/build/build_win.R
new file mode 100644
index 0000000000000000000000000000000000000000..e6861ceb5e94157a4ed21359a4d3339b9f1de8fb
--- /dev/null
+++ b/radiant.basics/build/build_win.R
@@ -0,0 +1,26 @@
+## build for windows
+rv <- R.Version()
+rv <- paste(rv$major, substr(rv$minor, 1, 1), sep = ".")
+
+rvprompt <- readline(prompt = paste0("Running for R version: ", rv, ". Is that what you wanted y/n: "))
+if (grepl("[nN]", rvprompt))
+ stop("Change R-version using Rstudio > Tools > Global Options > Rversion")
+
+## build for windows
+setwd(rstudioapi::getActiveProject())
+f <- devtools::build(binary = TRUE)
+devtools::install(upgrade = "never")
+
+fl <- list.files(pattern = "*.zip", path = "../", full.names = TRUE)
+
+for (f in fl) {
+ print(glue::glue("Copying: {f}"))
+ file.copy(f, "C:/Users/vnijs/Dropbox/r-packages/", overwrite = TRUE)
+ unlink(f)
+}
+
+#options(repos = c(RSM = "https://radiant-rstats.github.io/minicran"))
+#install.packages("radiant.data", type = "binary")
+# remove.packages(c("radiant.data", "radiant.model"))
+#install.packages("radiant.update")
+# radiant.update::radiant.update()
diff --git a/radiant.basics/data/consider.rda b/radiant.basics/data/consider.rda
new file mode 100644
index 0000000000000000000000000000000000000000..d00b31838f8bf9cbe39ca333d1fba82a47346ba9
Binary files /dev/null and b/radiant.basics/data/consider.rda differ
diff --git a/radiant.basics/data/demand_uk.rda b/radiant.basics/data/demand_uk.rda
new file mode 100644
index 0000000000000000000000000000000000000000..558d54557306b612a4faca6a794635d8e3ad3ce1
Binary files /dev/null and b/radiant.basics/data/demand_uk.rda differ
diff --git a/radiant.basics/data/newspaper.rda b/radiant.basics/data/newspaper.rda
new file mode 100644
index 0000000000000000000000000000000000000000..15cd23c5daf54ee27f71ac48adf1c6766017f220
Binary files /dev/null and b/radiant.basics/data/newspaper.rda differ
diff --git a/radiant.basics/data/salary.rda b/radiant.basics/data/salary.rda
new file mode 100644
index 0000000000000000000000000000000000000000..811a3223f58c686fb16683300103e95d6bad34b6
Binary files /dev/null and b/radiant.basics/data/salary.rda differ
diff --git a/radiant.basics/inst/app/global.R b/radiant.basics/inst/app/global.R
new file mode 100644
index 0000000000000000000000000000000000000000..f83f097571a1d7f92255ec4dcb32d6d5ebc00c5d
--- /dev/null
+++ b/radiant.basics/inst/app/global.R
@@ -0,0 +1,32 @@
+library(shiny.i18n)
+# file with translations
+i18n <- Translator$new(translation_csvs_path = "../translations")
+
+# change this to zh
+i18n$set_translation_language("zh")
+
+## sourcing from radiant.data
+options(radiant.path.data = system.file(package = "radiant.data"))
+source(file.path(getOption("radiant.path.data"), "app/global.R"), encoding = getOption("radiant.encoding", default = "UTF-8"), local = TRUE)
+
+ifelse(grepl("radiant.basics", getwd()) && file.exists("../../inst"), "..", system.file(package = "radiant.basics")) %>%
+ options(radiant.path.basics = .)
+
+## setting path for figures in help files
+addResourcePath("figures_basics", "tools/help/figures/")
+
+## setting path for www resources
+addResourcePath("www_basics", file.path(getOption("radiant.path.basics"), "app/www/"))
+
+## loading urls and ui
+source("init.R", encoding = getOption("radiant.encoding", "UTF-8"), local = TRUE)
+options(radiant.url.patterns = make_url_patterns())
+
+## if radiant.data is not in search main function from dplyr etc. won't be available
+if (!"package:radiant.basics" %in% search() &&
+ isTRUE(getOption("radiant.development")) &&
+ getOption("radiant.path.basics") == "..") {
+ options(radiant.from.package = FALSE)
+} else {
+ options(radiant.from.package = TRUE)
+}
diff --git a/radiant.basics/inst/app/help.R b/radiant.basics/inst/app/help.R
new file mode 100644
index 0000000000000000000000000000000000000000..040ebd73f3781357465085386964580480ed6beb
--- /dev/null
+++ b/radiant.basics/inst/app/help.R
@@ -0,0 +1,27 @@
+help_basics <- c(
+ "Probability calculator" = "prob_calc.md", "Central limit theorem" = "clt.md",
+ "Single mean" = "single_mean.md", "Compare means" = "compare_means.md",
+ "Single proportion" = "single_prop.md", "Compare proportions" = "compare_props.md",
+ "Goodness of fit" = "goodness.md", "Cross-tabs" = "cross_tabs.md",
+ "Correlation" = "correlation.md"
+)
+
+output$help_basics <- reactive(append_help("help_basics", file.path(getOption("radiant.path.basics"), "app/tools/help"), Rmd = TRUE))
+
+observeEvent(input$help_basics_all, {
+ help_switch(input$help_basics_all, "help_basics")
+})
+observeEvent(input$help_basics_none, {
+ help_switch(input$help_basics_none, "help_basics", help_on = FALSE)
+})
+
+help_basics_panel <- tagList(
+ wellPanel(
+ HTML(""),
+ checkboxGroupInput(
+ "help_basics", NULL, help_basics,
+ selected = state_group("help_basics"), inline = TRUE
+ )
+ )
+)
diff --git a/radiant.basics/inst/app/init.R b/radiant.basics/inst/app/init.R
new file mode 100644
index 0000000000000000000000000000000000000000..5f9513350f94f8488166992b6fbf5e26204ab920
--- /dev/null
+++ b/radiant.basics/inst/app/init.R
@@ -0,0 +1,49 @@
+## urls for menu
+r_url_list <- getOption("radiant.url.list")
+r_url_list[["Single mean"]] <-
+ list("tabs_single_mean" = list("Summary" = "basics/single-mean/", "Plot" = "basics/single-mean/plot/"))
+r_url_list[["Compare means"]] <-
+ list("tabs_compare_means" = list("Summary" = "basics/compare-means/", "Plot" = "basics/compare-means/plot/"))
+r_url_list[["Single proportion"]] <-
+ list("tabs_single_prop" = list("Summary" = "basics/single-prop/", "Plot" = "basics/single-prop/plot/"))
+r_url_list[["Compare proportions"]] <-
+ list("tabs_compare_props" = list("Summary" = "basics/compare-props/", "Plot" = "basics/compare-props/plot/"))
+r_url_list[["Goodness of fit"]] <-
+ list("tabs_goodness" = list("Summary" = "basics/goodness/", "Plot" = "basics/goodness/plot/"))
+r_url_list[["Cross-tabs"]] <-
+ list("tabs_cross_tabs" = list("Summary" = "basics/cross-tabs/", "Plot" = "basics/cross-tabs/plot/"))
+r_url_list[["Correlation"]] <-
+ list("tabs_correlation" = list("Summary" = "basics/correlation/", "Plot" = "basics`/correlation/plot/"))
+options(radiant.url.list = r_url_list)
+rm(r_url_list)
+
+## try http://127.0.0.1:3174/?url=basics/goodness/plot/&SSUID=local
+## http://127.0.0.1:7407/?url=basics/compare-means/plot/&SSUID=local-a82049
+
+## design menu
+options(
+ radiant.basics_ui =
+ tagList(
+ navbarMenu(
+ i18n$t("Basics"),
+ tags$head(
+ tags$script(src = "www_basics/js/run_return.js")
+ ),
+ i18n$t("Probability"),
+ tabPanel(i18n$t("Probability calculator"), uiOutput("prob_calc")),
+ tabPanel(i18n$t("Central Limit Theorem"), uiOutput("clt")),
+ "----", i18n$t("Means"),
+ tabPanel(i18n$t("Single mean"), uiOutput("single_mean")),
+ tabPanel(i18n$t("Compare means"), uiOutput("compare_means")),
+ tabPanel(i18n$t("Normality test"),uiOutput("normality_test")),
+ tabPanel(i18n$t("Homogeneity of variance test"),uiOutput("homo_variance_test")),
+ "----", i18n$t("Proportions"),
+ tabPanel(i18n$t("Single proportion"), uiOutput("single_prop")),
+ tabPanel(i18n$t("Compare proportions"), uiOutput("compare_props")),
+ "----", i18n$t("Tables"),
+ tabPanel(i18n$t("Goodness of fit"), uiOutput("goodness")),
+ tabPanel(i18n$t("Cross-tabs"), uiOutput("cross_tabs")),
+ tabPanel(i18n$t("Correlation"), uiOutput("correlation"))
+ )
+ )
+)
diff --git a/radiant.basics/inst/app/server.R b/radiant.basics/inst/app/server.R
new file mode 100644
index 0000000000000000000000000000000000000000..dd26d78c10b5e97367b84154896f2473ce022a9a
--- /dev/null
+++ b/radiant.basics/inst/app/server.R
@@ -0,0 +1,59 @@
+if (isTRUE(getOption("radiant.from.package"))) {
+ library(radiant.basics)
+}
+
+shinyServer(function(input, output, session) {
+
+ ## source shared functions
+ source(file.path(getOption("radiant.path.data"), "app/init.R"), encoding = getOption("radiant.encoding"), local = TRUE)
+ source(file.path(getOption("radiant.path.data"), "app/radiant.R"), encoding = getOption("radiant.encoding"), local = TRUE)
+ source("help.R", encoding = getOption("radiant.encoding"), local = TRUE)
+
+ ## help ui
+ output$help_basics_ui <- renderUI({
+ sidebarLayout(
+ sidebarPanel(
+ help_data_panel,
+ help_basics_panel,
+ uiOutput("help_text"),
+ width = 3
+ ),
+ mainPanel(
+ HTML(paste0("Select help files to show and search
")), + htmlOutput("help_data"), + htmlOutput("help_basics") + ) + ) + }) + + ## packages to use for example data + options(radiant.example.data = c("radiant.data", "radiant.basics")) + + ## source data & app tools from radiant.data + for (file in list.files( + c( + file.path(getOption("radiant.path.data"), "app/tools/app"), + file.path(getOption("radiant.path.data"), "app/tools/data") + ), + pattern = "\\.(r|R)$", full.names = TRUE + )) + source(file, encoding = getOption("radiant.encoding"), local = TRUE) + + ## 'sourcing' package functions in the server.R environment for development + if (!isTRUE(getOption("radiant.from.package"))) { + for (file in list.files("../../R", pattern = "\\.(r|R)$", full.names = TRUE)) { + source(file, encoding = getOption("radiant.encoding"), local = TRUE) + } + cat("\nGetting radiant.basics from source ...\n") + } else { + ## weired but required + summary.correlation <- radiant.basics:::summary.correlation + } + + ## source analysis tools for basic app + for (file in list.files(c("tools/analysis"), pattern = "\\.(r|R)$", full.names = TRUE)) + source(file, encoding = getOption("radiant.encoding"), local = TRUE) + + ## save state on refresh or browser close + saveStateOnRefresh(session) +}) diff --git a/radiant.basics/inst/app/tools/analysis/clt_ui.R b/radiant.basics/inst/app/tools/analysis/clt_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..fd2b94b3e8fb244ba6487f81e037c202a36ac251 --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/clt_ui.R @@ -0,0 +1,252 @@ +############################### +# Central Limit Theorem +############################### +clt_dist <- c("Normal", "Binomial", "Uniform", "Exponential") %>% + setNames(c( + i18n$t("Normal"), + i18n$t("Binomial"), + i18n$t("Uniform"), + i18n$t("Exponential") + )) + +clt_stat <- c("sum", "mean") %>% + setNames(c( + i18n$t("Sum"), + i18n$t("Mean") + )) +clt_args <- as.list(formals(clt)) + +clt_inputs <- reactive({ + for (i in names(clt_args)) { + clt_args[[i]] <- input[[paste0("clt_", i)]] + } + clt_args +}) + +## add a spinning refresh icon if the tabel needs to be (re)calculated +run_refresh(clt_args, "clt", init = "dist", label = i18n$t("Run simulation"), relabel = i18n$t("Re-run simulation"), data = FALSE) + +output$ui_clt <- renderUI({ + tagList( + wellPanel( + actionButton("clt_run", i18n$t("Run simulation"), width = "100%", icon = icon("play", verify_fa = FALSE), class = "btn-success") + ), + wellPanel( + selectInput( + "clt_dist", i18n$t("Distribution:"), + choices = clt_dist, + selected = state_single("clt_dist", clt_dist), + multiple = FALSE + ), + conditionalPanel( + condition = "input.clt_dist == 'Uniform'", + make_side_by_side( + numericInput( + "clt_unif_min", i18n$t("Min:"), + value = state_init("clt_unif_min", 0) + ), + numericInput( + "clt_unif_max", i18n$t("Max:"), + value = state_init("clt_unif_max", 1) + ) + ) + ), + conditionalPanel( + condition = "input.clt_dist == 'Normal'", + make_side_by_side( + numericInput( + "clt_norm_mean", i18n$t("Mean:"), + value = state_init("clt_norm_mean", 0) + ), + numericInput( + "clt_norm_sd", i18n$t("SD:"), + value = state_init("clt_norm_sd", 1), + min = 0.1, step = 0.1 + ) + ) + ), + conditionalPanel( + condition = "input.clt_dist == 'Exponential'", + numericInput( + "clt_expo_rate", i18n$t("Rate:"), + value = state_init("clt_expo_rate", 1), + min = 1, step = 1 + ) + ), + conditionalPanel( + condition = "input.clt_dist == 'Binomial'", + make_side_by_side( + numericInput( + "clt_binom_size", i18n$t("Size:"), + value = state_init("clt_binom_size", 10), + min = 1, step = 1 + ), + numericInput( + "clt_binom_prob", i18n$t("Prob:"), + value = state_init("clt_binom_prob", 0.2), + min = 0, max = 1, step = .1 + ) + ) + ), + make_side_by_side( + numericInput( + "clt_n", i18n$t("Sample size:"), + value = state_init("clt_n", 100), + min = 2, step = 1 + ), + numericInput( + "clt_m", i18n$t("# of samples:"), + value = state_init("clt_m", 100), + min = 2, step = 1 + ) + ), + sliderInput( + "clt_bins", + label = i18n$t("Number of bins:"), + min = 1, max = 50, step = 1, + value = state_init("clt_bins", 15), + ), + radioButtons( + "clt_stat", NULL, + choices = clt_stat, + selected = state_init("clt_stat", "sum"), + inline = TRUE + ) + ), + help_and_report( + modal_title = i18n$t("Central Limit Theorem"), fun_name = "clt", + help_file = inclRmd(file.path(getOption("radiant.path.basics"), "app/tools/help/clt.md")) + ) + ) +}) + +clt_plot_width <- function() 700 +clt_plot_height <- function() 700 + +## output is called from the main radiant ui.R +output$clt <- renderUI({ + register_plot_output( + "plot_clt", ".plot_clt", + height_fun = "clt_plot_height", + width_fun = "clt_plot_width" + ) + + ## two separate tabs + clt_output_panels <- tagList( + tabPanel( + "Plot", + download_link("dlp_clt"), + plotOutput("plot_clt", width = "100%", height = "100%") + ) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Probability"), + tool = i18n$t("Central Limit Theorem"), + data = NULL, + tool_ui = "ui_clt", + output_panels = clt_output_panels + ) +}) + +.clt <- eventReactive(input$clt_run, { + ## avoiding input errors + ret <- "" + if (is.na(input$clt_n) || input$clt_n < 2) { + ret <- i18n$t("Please choose a sample size larger than 2") + } else if (is.na(input$clt_m) || input$clt_m < 2) { + ret <- i18n$t("Please choose 2 or more samples") + } else if (input$clt_dist == "Uniform") { + if (is.na(input$clt_unif_min)) { + ret <- i18n$t("Please choose a minimum value for the uniform distribution") + } else if (is.na(input$clt_unif_max)) { + ret <- i18n$t("Please choose a maximum value for the uniform distribution") + } else if (input$clt_unif_max <= input$clt_unif_min) { + ret <- i18n$t("The maximum value for the uniform distribution\nmust be larger than the minimum value") + } + } else if (input$clt_dist == "Normal") { + if (is.na(input$clt_norm_mean)) { + ret <- i18n$t("Please choose a mean value for the normal distribution") + } else if (is.na(input$clt_norm_sd) || input$clt_norm_sd < .001) { + ret <- i18n$t("Please choose a non-zero standard deviation for the normal distribution") + } + } else if (input$clt_dist == "Exponential") { + if (is.na(input$clt_expo_rate) || input$clt_expo_rate < 1) { + ret <- i18n$t("Please choose a rate larger than 1 for the exponential distribution") + } + } else if (input$clt_dist == "Binomial") { + if (is.na(input$clt_binom_size) || input$clt_binom_size < 1) { + ret <- i18n$t("Please choose a size parameter larger than 1 for the binomial distribution") + } else if (is.na(input$clt_binom_prob) || input$clt_binom_prob < 0.01) { + ret <- i18n$t("Please choose a probability between 0 and 1 for the binomial distribution") + } + } + if (is.empty(ret)) { + do.call(clt, clt_inputs()) + } else { + ret + } +}) + +.plot_clt <- reactive({ + if (not_pressed(input$clt_run)) { + return(i18n$t("** Press the Run simulation button to simulate data **")) + } + clt <- .clt() + validate(need(!is.character(clt), paste0("\n\n\n ", clt))) + withProgress(message = i18n$t("Generating plots"), value = 1, { + plot(clt, stat = input$clt_stat, bins = input$clt_bins) + }) +}) + +clt_report <- function() { + outputs <- c("plot") + inp_out <- list(list(stat = input$clt_stat, bins = input$clt_bins)) + inp <- clt_inputs() + inp3 <- inp[!grepl("_", names(inp))] + if (input$clt_dist == "Normal") { + inp <- c(inp3, inp[grepl("norm_", names(inp))]) + } else if (input$clt_dist == "Uniform") { + inp <- c(inp3, inp[grepl("unif", names(inp))]) + } else if (input$clt_dist == "Binomial") { + inp <- c(inp3, inp[grepl("binom_", names(inp))]) + } else if (input$clt_dist == "Exponential") { + inp <- c(inp3, inp[grepl("expo_", names(inp))]) + } + + update_report( + inp_main = clean_args(inp, clt_args), + fun_name = "clt", + inp_out = inp_out, + outputs = outputs, + figs = TRUE, + fig.width = clt_plot_width(), + fig.height = clt_plot_height() + ) +} + +download_handler( + id = "dlp_clt", + fun = download_handler_plot, + fn = function() paste0(tolower(input$clt_dist), "_clt"), + type = "png", + caption = i18n$t("Save central limit theorem plot"), + plot = .plot_clt, + width = clt_plot_width, + height = clt_plot_height +) + +observeEvent(input$clt_report, { + r_info[["latest_screenshot"]] <- NULL + clt_report() +}) + +observeEvent(input$clt_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_clt_screenshot") +}) + +observeEvent(input$modal_clt_screenshot, { + clt_report() + removeModal() ## remove shiny modal after save +}) diff --git a/radiant.basics/inst/app/tools/analysis/compare_means_ui.R b/radiant.basics/inst/app/tools/analysis/compare_means_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..4e0de2e3ee302ed066b3c2507f65fc83205e9058 --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/compare_means_ui.R @@ -0,0 +1,317 @@ +## choice lists for compare means +cm_alt <- c( + "two.sided", + "less", + "greater" +) %>% setNames(c( + i18n$t("Two sided"), + i18n$t("Less than"), + i18n$t("Greater than") +)) + +cm_samples <- c( + "independent", + "paired" +) %>% setNames(c( + i18n$t("independent"), + i18n$t("paired") +)) + +cm_adjust <- c( + "none", + "bonf" +) %>% setNames(c( + i18n$t("None"), + i18n$t("Bonferroni") +)) + +cm_plots <- c( + "scatter", + "box", + "density", + "bar" +) %>% setNames(c( + i18n$t("Scatter"), + i18n$t("Box"), + i18n$t("Density"), + i18n$t("Bar") +)) +## list of function arguments +cm_args <- as.list(formals(compare_means)) + +## list of function inputs selected by user +cm_inputs <- reactive({ + ## loop needed because reactive values don't allow single bracket indexing + cm_args$data_filter <- if (input$show_filter) input$data_filter else "" + cm_args$dataset <- input$dataset + for (i in r_drop(names(cm_args))) { + cm_args[[i]] <- input[[paste0("cm_", i)]] + } + cm_args +}) + +############################### +# Compare means +############################### +output$ui_cm_var1 <- renderUI({ + vars <- c("None" = "", groupable_vars()) + isNum <- .get_class() %in% c("integer", "numeric", "ts") + + ## can't use unique here - removes variable type information + vars <- c(vars, varnames()[isNum]) %>% .[!duplicated(.)] + + selectInput( + inputId = "cm_var1", + label = i18n$t("Select a factor or numeric variable:"), + choices = vars, + selected = state_single("cm_var1", vars), + multiple = FALSE + ) +}) + +output$ui_cm_var2 <- renderUI({ + if (not_available(input$cm_var1)) { + return() + } + isNum <- .get_class() %in% c("integer", "numeric", "ts") + vars <- varnames()[isNum] + + if (input$cm_var1 %in% vars) { + ## when cm_var1 is numeric comparisons for multiple variables are possible + vars <- vars[-which(vars == input$cm_var1)] + if (length(vars) == 0) { + return() + } + + selectizeInput( + inputId = "cm_var2", label = i18n$t("Numeric variable(s):"), + selected = state_multiple("cm_var2", vars, isolate(input$cm_var2)), + choices = vars, multiple = TRUE, + options = list(placeholder = "None", plugins = list("remove_button", "drag_drop")) + ) + } else { + ## when cm_var1 is not numeric comparisons are across levels/groups + vars <- c("None" = "", vars) + selectInput( + "cm_var2", i18n$t("Numeric variable:"), + selected = state_single("cm_var2", vars), + choices = vars, + multiple = FALSE + ) + } +}) + +output$ui_cm_comb <- renderUI({ + if (not_available(input$cm_var1)) { + return() + } + + if (.get_class()[[input$cm_var1]] == "factor") { + levs <- .get_data()[[input$cm_var1]] %>% levels() + } else { + levs <- c(input$cm_var1, input$cm_var2) + } + + if (length(levs) > 2) { + cmb <- combn(levs, 2) %>% apply(2, paste, collapse = ":") + } else { + return() + } + + selectizeInput( + "cm_comb", + label = i18n$t("Choose combinations:"), + choices = cmb, + selected = state_multiple("cm_comb", cmb, cmb[1]), + multiple = TRUE, + options = list(placeholder = i18n$t("Evaluate all combinations"), plugins = list("remove_button", "drag_drop")) + ) +}) + + +output$ui_compare_means <- renderUI({ + req(input$dataset) + tagList( + wellPanel( + conditionalPanel( + uiOutput("ui_cm_var1"), + uiOutput("ui_cm_var2"), + condition = "input.tabs_compare_means == 'Summary'", + uiOutput("ui_cm_comb"), + selectInput( + inputId = "cm_alternative", label = i18n$t("Alternative hypothesis:"), + choices = cm_alt, + selected = state_single("cm_alternative", cm_alt, cm_args$alternative) + ), + sliderInput( + "cm_conf_lev", i18n$t("Confidence level:"), + min = 0.85, max = 0.99, step = 0.01, + value = state_init("cm_conf_lev", cm_args$conf_lev) + ), + checkboxInput("cm_show", i18n$t("Show additional statistics"), value = state_init("cm_show", FALSE)), + radioButtons( + inputId = "cm_samples", label = i18n$t("Sample type:"), cm_samples, + selected = state_init("cm_samples", cm_args$samples), + inline = TRUE + ), + radioButtons( + inputId = "cm_adjust", label = i18n$t("Multiple comp. adjustment:"), cm_adjust, + selected = state_init("cm_adjust", cm_args$adjust), + inline = TRUE + ), + radioButtons( + inputId = "cm_test", label = i18n$t("Test type:"), + choices = c( + "t", + "wilcox" + ) %>% setNames(c( + i18n$t("t-test"), + i18n$t("Wilcox") + )), + selected = state_init("cm_test", cm_args$test), + inline = TRUE + ) + ), + conditionalPanel( + condition = "input.tabs_compare_means == 'Plot'", + selectizeInput( + inputId = "cm_plots", label = i18n$t("Select plots:"), + choices = cm_plots, + selected = state_multiple("cm_plots", cm_plots, "scatter"), + multiple = TRUE, + options = list(placeholder = i18n$t("Select plots"), plugins = list("remove_button", "drag_drop")) + ) + ) + ), + help_and_report( + modal_title = i18n$t("Compare means"), + fun_name = "compare_means", + help_file = inclMD(file.path(getOption("radiant.path.basics"), "app/tools/help/compare_means.md")) + ) + ) +}) + +cm_plot <- reactive({ + list(plot_width = 650, plot_height = 400 * max(length(input$cm_plots), 1)) +}) + +cm_plot_width <- function() { + cm_plot() %>% + (function(x) if (is.list(x)) x$plot_width else 650) +} + +cm_plot_height <- function() { + cm_plot() %>% + (function(x) if (is.list(x)) x$plot_height else 400) +} + +# output is called from the main radiant ui.R +output$compare_means <- renderUI({ + register_print_output("summary_compare_means", ".summary_compare_means", ) + register_plot_output( + "plot_compare_means", ".plot_compare_means", + height_fun = "cm_plot_height" + ) + + # two separate tabs + cm_output_panels <- tabsetPanel( + id = "tabs_compare_means", + tabPanel(i18n$t("Summary"), value = "Summary", verbatimTextOutput("summary_compare_means")), + tabPanel( + i18n$t("Plot"), value = "Plot", + download_link("dlp_compare_means"), + plotOutput("plot_compare_means", height = "100%") + ) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Means"), + tool = i18n$t("Compare means"), + tool_ui = "ui_compare_means", + output_panels = cm_output_panels + ) +}) + +cm_available <- reactive({ + if (not_available(input$cm_var1) || not_available(input$cm_var2)) { + return(i18n$t("This analysis requires at least two variables. The first can be of type\nfactor, numeric, or interval. The second must be of type numeric or interval.\nIf these variable types are not available please select another dataset.\n\n") %>% suggest_data("salary")) + } else if (length(input$cm_var2) > 1 && .get_class()[input$cm_var1] == "factor") { + " " + } else if (input$cm_var1 %in% input$cm_var2) { + " " + } else { + "available" + } +}) + +.compare_means <- reactive({ + cmi <- cm_inputs() + cmi$envir <- r_data + do.call(compare_means, cmi) +}) + +.summary_compare_means <- reactive({ + if (cm_available() != "available") { + return(cm_available()) + } + if (input$cm_show) summary(.compare_means(), show = TRUE) else summary(.compare_means()) +}) + +.plot_compare_means <- reactive({ + if (cm_available() != "available") { + return(cm_available()) + } + validate(need(input$cm_plots, i18n$t("Nothing to plot. Please select a plot type"))) + withProgress(message = i18n$t("Generating plots"), value = 1, { + plot(.compare_means(), plots = input$cm_plots, shiny = TRUE) + }) +}) + +compare_means_report <- function() { + if (is.empty(input$cm_var1) || is.empty(input$cm_var2)) { + return(invisible()) + } + figs <- FALSE + outputs <- c("summary") + inp_out <- list(list(show = input$cm_show), "") + if (length(input$cm_plots) > 0) { + outputs <- c("summary", "plot") + inp_out[[2]] <- list(plots = input$cm_plots, custom = FALSE) + figs <- TRUE + } + update_report( + inp_main = clean_args(cm_inputs(), cm_args), + fun_name = "compare_means", + inp_out = inp_out, + outputs = outputs, + figs = figs, + fig.width = cm_plot_width(), + fig.height = cm_plot_height() + ) +} + +download_handler( + id = "dlp_compare_means", + fun = download_handler_plot, + fn = function() paste0(input$dataset, "_compare_means"), + type = "png", + caption = i18n$t("Save compare means plot"), + plot = .plot_compare_means, + width = cm_plot_width, + height = cm_plot_height +) + +observeEvent(input$compare_means_report, { + r_info[["latest_screenshot"]] <- NULL + compare_means_report() +}) + +observeEvent(input$compare_means_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_compare_means_screenshot") +}) + +observeEvent(input$modal_compare_means_screenshot, { + compare_means_report() + removeModal() ## remove shiny modal after save +}) diff --git a/radiant.basics/inst/app/tools/analysis/compare_props_ui.R b/radiant.basics/inst/app/tools/analysis/compare_props_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..8e814a8968b3798b2708305b2990c5dae1614803 --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/compare_props_ui.R @@ -0,0 +1,281 @@ +## choice lists for compare proportions(不使用等号命名) +cp_alt <- c("two.sided", "less", "greater") %>% + setNames(c( + i18n$t("Two sided"), + i18n$t("Less than"), + i18n$t("Greater than") + )) + +cp_adjust <- c("none", "bonf") %>% + setNames(c( + i18n$t("None"), + i18n$t("Bonferroni") + )) + +# cp_plots <- c("props", "counts") %>% setNames(c(i18n$t("Proportions"), i18n$t("Relative"))) +cp_plots <- c("bar", "dodge") %>% + setNames(c( + i18n$t("Bar"), + i18n$t("Dodge") + )) + +## list of function arguments +cp_args <- as.list(formals(compare_props)) + +## list of function inputs selected by user +cp_inputs <- reactive({ + ## loop needed because reactive values don't allow single bracket indexing + cp_args$data_filter <- if (input$show_filter) input$data_filter else "" + cp_args$dataset <- input$dataset + for (i in r_drop(names(cp_args))) { + cp_args[[i]] <- input[[paste0("cp_", i)]] + } + cp_args +}) + +############################### +# Compare proportions +############################### +output$ui_cp_var1 <- renderUI({ + vars <- c("None" = "", groupable_vars()) + selectInput( + "cp_var1", i18n$t("Select a grouping variable:"), + choices = vars, + selected = state_single("cp_var1", vars), + multiple = FALSE + ) +}) + +output$ui_cp_var2 <- renderUI({ + vars <- two_level_vars() + if (not_available(input$cp_var1)) { + return() + } + if (input$cp_var1 %in% vars) vars <- vars[-which(vars == input$cp_var1)] + + vars <- c("None" = "", vars) + selectInput( + inputId = "cp_var2", i18n$t("Variable (select one):"), + selected = state_single("cp_var2", vars), + choices = vars, + multiple = FALSE + ) +}) + +output$ui_cp_levs <- renderUI({ + if (not_available(input$cp_var2)) { + return() + } else { + levs <- .get_data()[[input$cp_var2]] %>% + as.factor() %>% + levels() + } + + selectInput( + inputId = "cp_levs", i18n$t("Choose level:"), + choices = levs, + selected = state_single("cp_levs", levs), + multiple = FALSE + ) +}) + +output$ui_cp_comb <- renderUI({ + if (not_available(input$cp_var1)) { + return() + } + + dat <- .get_data()[[input$cp_var1]] %>% as.factor() + levs <- levels(dat) + alevs <- unique(dat) + len <- length(dat) + levs <- levs[levs %in% alevs] + + if (length(levs) > 2 && length(levs) < len) { + cmb <- combn(levs, 2) %>% apply(2, paste, collapse = ":") + } else { + return() + } + + selectizeInput( + "cp_comb", i18n$t("Choose combinations:"), + choices = cmb, + selected = state_multiple("cp_comb", cmb, cmb[1]), + multiple = TRUE, + options = list(placeholder = i18n$t("Evaluate all combinations"), plugins = list("remove_button", "drag_drop")) + ) +}) + + +output$ui_compare_props <- renderUI({ + req(input$dataset) + tagList( + wellPanel( + conditionalPanel( + condition = "input.tabs_compare_props == 'Summary'", + uiOutput("ui_cp_var1"), + uiOutput("ui_cp_var2"), + uiOutput("ui_cp_levs"), + uiOutput("ui_cp_comb"), + selectInput( + inputId = "cp_alternative", i18n$t("Alternative hypothesis:"), + choices = cp_alt, + selected = state_single("cp_alternative", cp_alt, cp_args$alternative) + ), + checkboxInput( + "cp_show", i18n$t("Show additional statistics"), + value = state_init("cp_show", FALSE) + ), + sliderInput( + "cp_conf_lev", i18n$t("Confidence level:"), + min = 0.85, max = 0.99, step = 0.01, + value = state_init("cp_conf_lev", cp_args$conf_lev) + ), + radioButtons( + inputId = "cp_adjust", i18n$t("Multiple comp. adjustment:"), + cp_adjust, + selected = state_init("cp_adjust", cp_args$adjust), + inline = TRUE + ) + ), + conditionalPanel( + condition = "input.tabs_compare_props == 'Plot'", + selectizeInput( + inputId = "cp_plots", label = i18n$t("Select plots:"), + choices = cp_plots, + selected = state_multiple("cp_plots", cp_plots, "bar"), + multiple = TRUE, + options = list(placeholder = i18n$t("Select plots"), plugins = list("remove_button", "drag_drop")) + ) + ) + ), + help_and_report( + modal_title = i18n$t("Compare proportions"), + fun_name = "compare_props", + help_file = inclMD(file.path(getOption("radiant.path.basics"), "app/tools/help/compare_props.md")) + ) + ) +}) + +cp_plot <- reactive({ + list(plot_width = 650, plot_height = 400 * max(length(input$cp_plots), 1)) +}) + +cp_plot_width <- function() { + cp_plot() %>% + (function(x) if (is.list(x)) x$plot_width else 650) +} + +cp_plot_height <- function() { + cp_plot() %>% + (function(x) if (is.list(x)) x$plot_height else 400) +} + +# output is called from the main radiant ui.R +output$compare_props <- renderUI({ + register_print_output("summary_compare_props", ".summary_compare_props", ) + register_plot_output( + "plot_compare_props", ".plot_compare_props", + height_fun = "cp_plot_height" + ) + + # two separate tabs + cp_output_panels <- tabsetPanel( + id = "tabs_compare_props", + tabPanel(i18n$t("Summary"), value = "Summary", verbatimTextOutput("summary_compare_props")), + tabPanel( + i18n$t("Plot"), value = "Plot", + download_link("dlp_compare_props"), + plotOutput("plot_compare_props", height = "100%") + ) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Proportions"), + tool = i18n$t("Compare proportions"), + tool_ui = "ui_compare_props", + output_panels = cp_output_panels + ) +}) + +cp_available <- reactive({ + if (not_available(input$cp_var1) || not_available(input$cp_var2)) { + i18n$t("This analysis requires two categorical variables. The first must have\ntwo or more levels. The second can have only two levels. If these\nvariable types are not available please select another dataset.\n\n") %>% suggest_data("titanic") + } else if (input$cp_var1 %in% input$cp_var2) { + " " + } else { + "available" + } +}) + +.compare_props <- reactive({ + cpi <- cp_inputs() + cpi$envir <- r_data + do.call(compare_props, cpi) +}) + +.summary_compare_props <- reactive({ + if (cp_available() != "available") { + return(cp_available()) + } + if (input$cp_show) summary(.compare_props(), show = TRUE) else summary(.compare_props()) +}) + +.plot_compare_props <- reactive({ + if (cp_available() != "available") { + return(cp_available()) + } + validate(need(input$cp_plots, i18n$t("Nothing to plot. Please select a plot type"))) + withProgress(message = i18n$t("Generating plots"), value = 1, { + plot(.compare_props(), plots = input$cp_plots, shiny = TRUE) + }) +}) + +compare_props_report <- function() { + if (is.empty(input$cp_var1) || is.empty(input$cp_var2)) { + return(invisible()) + } + figs <- FALSE + outputs <- c("summary") + inp_out <- list(list(show = input$cp_show), "") + if (length(input$cp_plots) > 0) { + outputs <- c("summary", "plot") + inp_out[[2]] <- list(plots = input$cp_plots, custom = FALSE) + figs <- TRUE + } + + update_report( + inp_main = clean_args(cp_inputs(), cp_args), + fun_name = "compare_props", + inp_out = inp_out, + outputs = outputs, + figs = figs, + fig.width = cp_plot_width(), + fig.height = cp_plot_height() + ) +} + +download_handler( + id = "dlp_compare_props", + fun = download_handler_plot, + fn = function() paste0(input$dataset, "_compare_props"), + type = "png", + caption = i18n$t("Save compare proportions plot"), + plot = .plot_compare_props, + width = cp_plot_width, + height = cp_plot_height +) + +observeEvent(input$compare_props_report, { + r_info[["latest_screenshot"]] <- NULL + compare_props_report() +}) + +observeEvent(input$compare_props_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_compare_props_screenshot") +}) + +observeEvent(input$modal_compare_props_screenshot, { + compare_props_report() + removeModal() ## remove shiny modal after save +}) diff --git a/radiant.basics/inst/app/tools/analysis/correlation_ui.R b/radiant.basics/inst/app/tools/analysis/correlation_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..89417d747800f92868710e3152439645f13230d5 --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/correlation_ui.R @@ -0,0 +1,330 @@ +############################### +## Correlation +############################### +cor_method <- c( + "pearson", + "spearman", + "kendall" +) %>% setNames(c( + i18n$t("Pearson"), + i18n$t("Spearman"), + i18n$t("Kendall") +)) +## list of function arguments +cor_args <- as.list(formals(correlation)) + +## list of function inputs selected by user +cor_inputs <- reactive({ + ## loop needed because reactive values don't allow single bracket indexing + cor_args$data_filter <- if (input$show_filter) input$data_filter else "" + cor_args$dataset <- input$dataset + for (i in r_drop(names(cor_args))) { + cor_args[[i]] <- input[[paste0("cor_", i)]] + } + cor_args +}) + +output$ui_cor_method <- renderUI({ + if (isTRUE(input$cor_hcor)) { + cor_method <- c("pearson") %>% + setNames(c(i18n$t("Pearson"))) + } + selectInput( + "cor_method", i18n$t("Method:"), + choices = cor_method, + selected = state_single("cor_method", cor_method, "pearson"), + multiple = FALSE + ) +}) + +cor_sum_args <- as.list(if (exists("summary.correlation")) { + formals(summary.correlation) +} else { + formals(radiant.basics::summary.correlation) +}) + +## list of function inputs selected by user +cor_sum_inputs <- reactive({ + ## loop needed because reactive values don't allow single bracket indexing + for (i in names(cor_sum_args)) { + cor_sum_args[[i]] <- input[[paste0("cor_", i)]] + } + cor_sum_args +}) + +output$ui_cor_vars <- renderUI({ + withProgress(message = i18n$t("Acquiring variable information"), value = 1, { + vars <- varnames() + toSelect <- .get_class() %in% c("numeric", "integer", "date", "factor") + vars <- vars[toSelect] + }) + if (length(vars) == 0) { + return() + } + selectInput( + inputId = "cor_vars", label = i18n$t("Select variables:"), + choices = vars, + selected = state_multiple("cor_vars", vars, isolate(input$cor_vars)), + multiple = TRUE, + size = min(10, length(vars)), + selectize = FALSE + ) +}) + +output$ui_cor_nrobs <- renderUI({ + nrobs <- nrow(.get_data()) + choices <- c("1,000" = 1000, "5,000" = 5000, "10,000" = 10000, "All" = -1) %>% + .[. < nrobs] + selectInput( + "cor_nrobs", i18n$t("Number of data points plotted:"), + choices = choices, + selected = state_single("cor_nrobs", choices, 1000) + ) +}) + +output$ui_cor_name <- renderUI({ + req(input$dataset) + textInput("cor_name", i18n$t("Store as data.frame:"), "", placeholder = "Provide a name") +}) + +## add a spinning refresh icon if correlations need to be (re)calculated +run_refresh(cor_args, "cor", init = "vars", tabs = "tabs_correlation", label = i18n$t("Calculate correlation"), relabel = i18n$t("Re-calculate correlations")) + +output$ui_correlation <- renderUI({ + req(input$dataset) + tagList( + conditionalPanel( + condition = "input.tabs_correlation == 'Summary'", + wellPanel( + actionButton("cor_run", i18n$t("Calculate correlation"), width = "100%", icon = icon("play", verify_fa = FALSE), class = "btn-success") + ) + ), + wellPanel( + conditionalPanel( + condition = "input.tabs_correlation == 'Summary'", + uiOutput("ui_cor_vars"), + uiOutput("ui_cor_method"), + checkboxInput("cor_hcor", i18n$t("Adjust for {factor} variables"), value = state_init("cor_hcor", FALSE)), + conditionalPanel( + condition = "input.cor_hcor == true", + checkboxInput("cor_hcor_se", i18n$t("Calculate adjusted p.values"), value = state_init("cor_hcor_se", FALSE)) + ), + numericInput( + "cor_cutoff", i18n$t("Correlation cutoff:"), + min = 0, max = 1, step = 0.05, + value = state_init("cor_cutoff", 0) + ), + conditionalPanel( + condition = "input.cor_hcor == false", + checkboxInput( + "cor_covar", i18n$t("Show covariance matrix"), + value = state_init("cor_covar", FALSE) + ) + ), + ), + conditionalPanel( + condition = "input.tabs_correlation == 'Plot'", + uiOutput("ui_cor_nrobs") + ) + ), + conditionalPanel( + condition = "input.tabs_correlation == 'Summary'", + wellPanel( + tags$table( + tags$td(uiOutput("ui_cor_name")), + tags$td(actionButton("cor_store", i18n$t("Store"), icon = icon("plus", verify_fa = FALSE)), class = "top") + ) + ) + ), + help_and_report( + modal_title = i18n$t("Correlation"), + fun_name = "correlation", + help_file = inclMD(file.path(getOption("radiant.path.basics"), "app/tools/help/correlation.md")) + ) + ) +}) + +observeEvent(input$cor_hcor, { + if (input$cor_hcor == FALSE) { + updateCheckboxInput(session, "cor_hcor_se", value = FALSE) + } else { + updateCheckboxInput(session, "cor_covar", value = FALSE) + } +}) + +cor_plot <- reactive({ + max(2, length(input$cor_vars)) %>% + (function(x) list(plot_width = 400 + 75 * x, plot_height = 400 + 75 * x)) +}) + +cor_plot_width <- function() { + cor_plot() %>% + (function(x) if (is.list(x)) x$plot_width else 650) +} + +cor_plot_height <- function() { + cor_plot() %>% + (function(x) if (is.list(x)) x$plot_height else 650) +} + +## output is called from the main radiant ui.R +output$correlation <- renderUI({ + register_print_output("summary_correlation", ".summary_correlation") + register_plot_output( + "plot_correlation", ".plot_correlation", + height_fun = "cor_plot_height", + width_fun = "cor_plot_width" + ) + + ## two separate tabs + cor_output_panels <- tabsetPanel( + id = "tabs_correlation", + tabPanel(i18n$t("Summary"), value = "Summary", verbatimTextOutput("summary_correlation")), + tabPanel( + i18n$t("Plot"), value = "Plot", + download_link("dlp_correlation"), + plotOutput( + "plot_correlation", + width = "100%", + height = "100%" + ) + ) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Tables"), + tool = i18n$t("Correlation"), + tool_ui = "ui_correlation", + output_panels = cor_output_panels + ) +}) + +cor_available <- reactive({ + if (not_available(input$cor_vars) || length(input$cor_vars) < 2) { + return(i18n$t("This analysis requires two or more variables or type numeric,\ninteger,or date. If these variable types are not available\nplease select another dataset.\n\n") %>% suggest_data("salary")) + } + "available" +}) + +# .correlation <- reactive({ +.correlation <- eventReactive(input$cor_run, { + cori <- cor_inputs() + cori$envir <- r_data + do.call(correlation, cori) +}) + +.summary_correlation <- reactive({ + if (cor_available() != "available") { + return(cor_available()) + } + if (not_pressed(input$cor_run)) { + return(i18n$t("** Press the Calculate correlation button to generate output **")) + } + validate( + need( + input$cor_cutoff >= 0 && input$cor_cutoff <= 1, + i18n$t("Provide a correlation cutoff value in the range from 0 to 1") + ) + ) + withProgress(message = i18n$t("Calculating correlations"), value = 0.5, { + do.call(summary, c(list(object = .correlation()), cor_sum_inputs())) + }) +}) + +.plot_correlation <- reactive({ + if (cor_available() != "available") { + return(cor_available()) + } + if (not_pressed(input$cor_run)) { + return(i18n$t("** Press the Calculate correlation button to generate output **")) + } + req(input$cor_nrobs) + withProgress(message = i18n$t("Generating correlation plot"), value = 0.5, { + capture_plot(plot(.correlation(), nrobs = input$cor_nrobs)) + }) +}) + +correlation_report <- function() { + if (length(input$cor_vars) < 2) { + return(invisible()) + } + inp_out <- list("", "") + nrobs <- ifelse(is.empty(input$cor_nrobs), 1000, as_integer(input$cor_nrobs)) + inp_out[[1]] <- clean_args(cor_sum_inputs(), cor_sum_args[-1]) + inp_out[[2]] <- list(nrobs = nrobs) + + if (!is.empty(input$cor_name)) { + dataset <- fix_names(input$cor_name) + if (input$cor_name != dataset) { + updateTextInput(session, inputId = "cor_name", value = dataset) + } + xcmd <- paste0(dataset, " <- cor2df(result)\nregister(\"", dataset, "\", descr = result$descr)") + } else { + xcmd <- "" + } + + update_report( + inp_main = clean_args(cor_inputs(), cor_args), + fun_name = "correlation", + inp_out = inp_out, + fig.width = cor_plot_width(), + fig.height = cor_plot_height(), + xcmd = xcmd + ) +} + +download_handler( + id = "dlp_correlation", + fun = download_handler_plot, + fn = function() paste0(input$dataset, "_correlation"), + type = "png", + caption = i18n$t("Save correlation plot"), + plot = .plot_correlation, + width = cor_plot_width, + height = cor_plot_height +) + +observeEvent(input$cor_store, { + req(input$cor_name) + cmat <- try(.correlation(), silent = TRUE) + if (inherits(cmat, "try-error") || is.null(cmat)) { + return() + } + + dataset <- fix_names(input$cor_name) + updateTextInput(session, inputId = "cor_name", value = dataset) + r_data[[dataset]] <- cor2df(cmat) + register(dataset, descr = cmat$descr) + updateSelectInput(session, "dataset", selected = input$dataset) + + ## See https://shiny.posit.co//reference/shiny/latest/modalDialog.html + showModal( + modalDialog( + title = i18n$t("Data Stored"), + span( + i18n$t( + "Dataset '{dataset}' was successfully added to the datasets dropdown. Add code to Report > Rmd or Report > R to (re)create the results by clicking the report icon on the bottom left of your screen.", + dataset = dataset + ) + ), + footer = modalButton(i18n$t("OK")), + size = "s", + easyClose = TRUE + ) + ) +}) + +observeEvent(input$correlation_report, { + r_info[["latest_screenshot"]] <- NULL + correlation_report() +}) + +observeEvent(input$correlation_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_correlation_screenshot") +}) + +observeEvent(input$modal_correlation_screenshot, { + correlation_report() + removeModal() ## remove shiny modal after save +}) diff --git a/radiant.basics/inst/app/tools/analysis/cross_tabs_ui.R b/radiant.basics/inst/app/tools/analysis/cross_tabs_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..b349d232b44e881711c0d8a8f91f0cee62284958 --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/cross_tabs_ui.R @@ -0,0 +1,211 @@ +## alternative hypothesis options +ct_check <- c( + "observed", + "expected", + "chi_sq", + "dev_std", + "row_perc", + "col_perc", + "perc" +) + +names(ct_check) <- c( + i18n$t("Observed"), + i18n$t("Expected"), + i18n$t("Chi-squared"), + i18n$t("Deviation std."), + i18n$t("Row percentages"), + i18n$t("Column percentages"), + i18n$t("Table percentages") +) + +## list of function arguments +ct_args <- as.list(formals(cross_tabs)) + +## list of function inputs selected by user +ct_inputs <- reactive({ + ## loop needed because reactive values don't allow single bracket indexing + ct_args$data_filter <- if (input$show_filter) input$data_filter else "" + ct_args$dataset <- input$dataset + for (i in r_drop(names(ct_args))) { + ct_args[[i]] <- input[[paste0("ct_", i)]] + } + ct_args +}) + +############################### +# Cross-tabs +############################### +output$ui_ct_var1 <- renderUI({ + vars <- c("None" = "", groupable_vars()) + selectInput( + inputId = "ct_var1", label = i18n$t("Select a categorical variable:"), + choices = vars, selected = state_single("ct_var1", vars), multiple = FALSE + ) +}) + +output$ui_ct_var2 <- renderUI({ + if (not_available(input$ct_var1)) { + return() + } + vars <- c("None" = "", groupable_vars()) + + if (length(vars) > 0) vars <- vars[-which(vars == input$ct_var1)] + selectInput( + inputId = "ct_var2", label = i18n$t("Select a categorical variable:"), + selected = state_single("ct_var2", vars), + choices = vars, multiple = FALSE + ) +}) + +output$ui_cross_tabs <- renderUI({ + req(input$dataset) + tagList( + wellPanel( + conditionalPanel( + condition = "input.tabs_cross_tabs == 'Summary'", + uiOutput("ui_ct_var1"), + uiOutput("ui_ct_var2") + ), + br(), + checkboxGroupInput( + "ct_check", NULL, + choices = ct_check, + selected = state_group("ct_check"), + inline = FALSE + ) + ), + help_and_report( + modal_title = i18n$t("Cross-tabs"), + fun_name = "cross_tabs", + help_file = inclMD(file.path(getOption("radiant.path.basics"), "app/tools/help/cross_tabs.md")) + ) + ) +}) + +ct_plot <- reactive({ + list(plot_width = 650, plot_height = 400 * max(length(input$ct_check), 1)) +}) + +ct_plot_width <- function() { + ct_plot() %>% + (function(x) if (is.list(x)) x$plot_width else 650) +} + +ct_plot_height <- function() { + ct_plot() %>% + (function(x) if (is.list(x)) x$plot_height else 400) +} + +## output is called from the main radiant ui.R +output$cross_tabs <- renderUI({ + register_print_output("summary_cross_tabs", ".summary_cross_tabs") + register_plot_output( + "plot_cross_tabs", ".plot_cross_tabs", + height_fun = "ct_plot_height", + width_fun = "ct_plot_width" + ) + + ## two separate tabs + ct_output_panels <- tabsetPanel( + id = "tabs_cross_tabs", + tabPanel(i18n$t("Summary"), value = "Summary", verbatimTextOutput("summary_cross_tabs")), + tabPanel( + i18n$t("Plot"), value = "Plot", + download_link("dlp_cross_tabs"), + plotOutput("plot_cross_tabs", width = "100%", height = "100%") + ) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Tables"), + tool = i18n$t("Cross-tabs"), + tool_ui = "ui_cross_tabs", + output_panels = ct_output_panels + ) +}) + +ct_available <- reactive({ + if (not_available(input$ct_var1) || not_available(input$ct_var2)) { + i18n$t("This analysis requires two categorical variables. Both must have two or more levels.\nIf these variable types are not available please select another dataset.\n\n") %>% + suggest_data("newspaper") + } else { + "available" + } +}) + +.cross_tabs <- reactive({ + cti <- ct_inputs() + cti$envir <- r_data + do.call(cross_tabs, cti) +}) + +.summary_cross_tabs <- reactive({ + if (ct_available() != "available") { + return(ct_available()) + } + summary(.cross_tabs(), check = input$ct_check) +}) + +.plot_cross_tabs <- reactive({ + if (ct_available() != "available") { + return(ct_available()) + } + validate(need(input$ct_check, i18n$t("Nothing to plot. Please select a plot type"))) + withProgress(message = i18n$t("Generating plots"), value = 1, { + plot(.cross_tabs(), check = input$ct_check, shiny = TRUE) + }) +}) + +cross_tabs_report <- function() { + if (is.empty(input$ct_var1) || is.empty(input$ct_var2)) { + return(invisible()) + } + inp_out <- list("", "") + if (length(input$ct_check) > 0) { + outputs <- c("summary", "plot") + inp_out[[1]] <- list(check = input$ct_check) + inp_out[[2]] <- list(check = input$ct_check, custom = FALSE) + figs <- TRUE + } else { + outputs <- "summary" + inp_out[[1]] <- list(check = "") + figs <- FALSE + } + + update_report( + inp_main = clean_args(ct_inputs(), ct_args), + inp_out = inp_out, + fun_name = "cross_tabs", + outputs = outputs, + figs = figs, + fig.width = ct_plot_width(), + fig.height = ct_plot_height() + ) +} + +download_handler( + id = "dlp_cross_tabs", + fun = download_handler_plot, + fn = function() paste0(input$dataset, "_cross_tabs"), + type = "png", + caption = i18n$t("Save cross-tabs plot"), + plot = .plot_cross_tabs, + width = ct_plot_width, + height = ct_plot_height +) + +observeEvent(input$cross_tabs_report, { + r_info[["latest_screenshot"]] <- NULL + cross_tabs_report() +}) + +observeEvent(input$cross_tabs_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_cross_tabs_screenshot") +}) + +observeEvent(input$modal_cross_tabs_screenshot, { + cross_tabs_report() + removeModal() ## remove shiny modal after save +}) diff --git a/radiant.basics/inst/app/tools/analysis/goodness_ui.R b/radiant.basics/inst/app/tools/analysis/goodness_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..66723f09e901e95a16d4585dfe7e99d9532b6dae --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/goodness_ui.R @@ -0,0 +1,197 @@ +## alternative hypothesis options +gd_check <- c("observed", "expected", "chi_sq", "dev_std") +names(gd_check) <- c( + i18n$t("Observed"), + i18n$t("Expected"), + i18n$t("Chi-squared"), + i18n$t("Deviation std.") +) + +## list of function arguments +gd_args <- as.list(formals(goodness)) + +## list of function inputs selected by user +gd_inputs <- reactive({ + ## loop needed because reactive values don't allow single bracket indexing + gd_args$data_filter <- if (input$show_filter) input$data_filter else "" + gd_args$dataset <- input$dataset + for (i in r_drop(names(gd_args))) { + gd_args[[i]] <- input[[paste0("gd_", i)]] + } + gd_args +}) + +############################### +# Goodness of fit test +############################### +output$ui_gd_var <- renderUI({ + vars <- c("None" = "", groupable_vars()) + selectInput( + "gd_var", i18n$t("Select a categorical variable:"), + choices = vars, + selected = state_single("gd_var", vars), + multiple = FALSE + ) +}) + +output$ui_gd_p <- renderUI({ + req(input$gd_var) + returnTextInput( + "gd_p", i18n$t("Probabilities:"), + value = state_init("gd_p", ""), + placeholder = i18n$t("Enter probabilities (e.g., 1/2 1/2)") + ) +}) + +output$ui_goodness <- renderUI({ + req(input$dataset) + tagList( + wellPanel( + conditionalPanel( + condition = "input.tabs_goodness == 'Summary'", + uiOutput("ui_gd_var"), + uiOutput("ui_gd_p"), + br() + ), + checkboxGroupInput( + "gd_check", NULL, + choices = gd_check, + selected = state_group("gd_check"), + inline = FALSE + ) + ), + help_and_report( + modal_title = i18n$t("Goodness of fit"), + fun_name = "goodness", + help_file = inclMD(file.path(getOption("radiant.path.basics"), "app/tools/help/goodness.md")) + ) + ) +}) + +gd_plot <- reactive({ + list(plot_width = 650, plot_height = 400 * max(length(input$gd_check), 1)) +}) + +gd_plot_width <- function() { + gd_plot() %>% + (function(x) if (is.list(x)) x$plot_width else 650) +} + +gd_plot_height <- function() { + gd_plot() %>% + (function(x) if (is.list(x)) x$plot_height else 400) +} + +## output is called from the main radiant ui.R +output$goodness <- renderUI({ + register_print_output("summary_goodness", ".summary_goodness") + register_plot_output( + "plot_goodness", ".plot_goodness", + height_fun = "gd_plot_height", + width_fun = "gd_plot_width" + ) + + ## two separate tabs + gd_output_panels <- tabsetPanel( + id = "tabs_goodness", + tabPanel(i18n$t("Summary"), value = "Summary", verbatimTextOutput("summary_goodness")), + tabPanel( + i18n$t("Plot"), value = "Plot", + download_link("dlp_goodness"), + plotOutput("plot_goodness", width = "100%", height = "100%") + ) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Tables"), + tool = i18n$t("Goodness of fit"), + tool_ui = "ui_goodness", + output_panels = gd_output_panels + ) +}) + +gd_available <- reactive({ + if (not_available(input$gd_var)) { + i18n$t("This analysis requires a categorical variables with two or more levels.\nIf such a variable type is not available please select another dataset.\n\n") %>% suggest_data("newspaper") + } else { + "available" + } +}) + +.goodness <- reactive({ + gdi <- gd_inputs() + gdi$envir <- r_data + do.call(goodness, gdi) +}) + +.summary_goodness <- reactive({ + if (gd_available() != "available") { + return(gd_available()) + } + summary(.goodness(), check = input$gd_check) +}) + +.plot_goodness <- reactive({ + if (gd_available() != "available") { + return(gd_available()) + } + validate(need(input$gd_check, i18n$t("Nothing to plot. Please select a plot type"))) + withProgress(message = i18n$t("Generating plots"), value = 1, { + plot(.goodness(), check = input$gd_check, shiny = TRUE) + }) +}) + +goodness_report <- function() { + if (is.empty(input$gd_var)) { + return(invisible()) + } + inp_out <- list("", "") + if (length(input$gd_check) > 0) { + outputs <- c("summary", "plot") + inp_out[[1]] <- list(check = input$gd_check) + inp_out[[2]] <- list(check = input$gd_check, custom = FALSE) + figs <- TRUE + } else { + outputs <- "summary" + figs <- FALSE + } + + gdi <- gd_inputs() + gdi$p <- radiant.data::make_vec(gdi$p) + + update_report( + inp_main = clean_args(gdi, gd_args), + inp_out = inp_out, + fun_name = "goodness", + outputs = outputs, + figs = figs, + fig.width = gd_plot_width(), + fig.height = gd_plot_height() + ) +} + +download_handler( + id = "dlp_goodness", + fun = download_handler_plot, + fn = function() paste0(input$dataset, "_goodness"), + type = "png", + caption = i18n$t("Save goodness of fit plot"), + plot = .plot_goodness, + width = gd_plot_width, + height = gd_plot_height +) + +observeEvent(input$goodness_report, { + r_info[["latest_screenshot"]] <- NULL + goodness_report() +}) + +observeEvent(input$goodness_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_goodness_screenshot") +}) + +observeEvent(input$modal_goodness_screenshot, { + goodness_report() + removeModal() ## remove shiny modal after save +}) diff --git a/radiant.basics/inst/app/tools/analysis/homo_variance_test_ui.R b/radiant.basics/inst/app/tools/analysis/homo_variance_test_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..7a400fd1c78ec4adb3dea18b99ac2c19f5d20d82 --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/homo_variance_test_ui.R @@ -0,0 +1,190 @@ +############################################ +## Homogeneity of variance test - ui +############################################ + +## 1. 翻译标签 +hv_method <- c("levene", "bartlett", "fligner") +names(hv_method) <- c(i18n$t("Levene"), + i18n$t("Bartlett"), + i18n$t("Fligner")) + +hv_plots <- c("hist", "density", "boxplot") +names(hv_plots) <- c(i18n$t("Histogram"), + i18n$t("Density"), + i18n$t("Boxplot")) + +## 2. 函数形参 +hv_args <- as.list(formals(homo_variance_test)) + +## 3. 收集输入 +hv_inputs <- reactive({ + hv_args$data_filter <- if (input$show_filter) input$data_filter else "" + hv_args$dataset <- input$dataset + for (i in r_drop(names(hv_args))) { + hv_args[[i]] <- input[[paste0("hv_", i)]] + } + hv_args +}) + +## 4. 变量选择(numeric + grouping) +output$ui_hv_var <- renderUI({ + isNum <- .get_class() %in% c("integer", "numeric", "ts") + vars <- c("None" = "", varnames()[isNum]) + selectInput( + inputId = "hv_var", label = i18n$t("Variable (select one):"), + choices = vars, selected = state_single("hv_var", vars), multiple = FALSE + ) +}) + +output$ui_hv_group <- renderUI({ + vars <- groupable_vars() + selectInput( + inputId = "hv_group", label = i18n$t("Grouping variable:"), + choices = vars, selected = state_single("hv_group", vars), multiple = FALSE + ) +}) + +## 5. 主 UI +output$ui_homo_variance_test <- renderUI({ + req(input$dataset) + tagList( + wellPanel( + conditionalPanel( + condition = "input.tabs_homo_variance_test == 'Summary'", + uiOutput("ui_hv_var"), + uiOutput("ui_hv_group"), + selectInput( + inputId = "hv_method", label = i18n$t("Test method:"), + choices = hv_method, + selected = state_single("hv_method", hv_method, "levene"), + multiple = FALSE + ), + sliderInput( + "hv_conf_lev", i18n$t("Confidence level:"), + min = 0.85, max = 0.99, + value = state_init("hv_conf_lev", 0.95), step = 0.01 + ) + ), + conditionalPanel( + condition = "input.tabs_homo_variance_test == 'Plot'", + selectizeInput( + inputId = "hv_plots", label = i18n$t("Select plots:"), + choices = hv_plots, + selected = state_multiple("hv_plots", hv_plots, "boxplot"), + multiple = TRUE, + options = list(placeholder = i18n$t("Select plots"), + plugins = list("remove_button", "drag_drop")) + ) + ) + ), + help_and_report( + modal_title = i18n$t("Homogeneity of variance test"), + fun_name = "homo_variance_test", + help_file = inclMD(file.path(getOption("radiant.path.basics"), + "app/tools/help/homo_variance_test.md")) + ) + ) +}) + +## 6. 画图尺寸 +hv_plot <- reactive({ + list(plot_width = 650, + plot_height = 400 * max(length(input$hv_plots), 1)) +}) +hv_plot_width <- function() hv_plot()$plot_width +hv_plot_height <- function() hv_plot()$plot_height + +## 7. 输出面板 +output$homo_variance_test <- renderUI({ + register_print_output("summary_homo_variance_test", ".summary_homo_variance_test") + register_plot_output("plot_homo_variance_test", ".plot_homo_variance_test", + height_fun = "hv_plot_height") + + hv_output_panels <- tabsetPanel( + id = "tabs_homo_variance_test", + tabPanel(title = i18n$t("Summary"), + value = "Summary", + verbatimTextOutput("summary_homo_variance_test")), + tabPanel(title = i18n$t("Plot"), + value = "Plot", + download_link("dlp_homo_variance_test"), + plotOutput("plot_homo_variance_test", height = "100%")) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Homogeneity"), + tool = i18n$t("Homogeneity of variance test"), + tool_ui = "ui_homo_variance_test", + output_panels = hv_output_panels + ) +}) + +## 8. 可用性检查 +hv_available <- reactive({ + if (not_available(input$hv_var)) + return(i18n$t("This analysis requires a numeric variable. If none are\navailable please select another dataset.") %>% suggest_data("demand_uk")) + if (not_available(input$hv_group)) + return(i18n$t("Please select a grouping variable.")) + "available" +}) + +## 9. 计算核心 +.homo_variance_test <- reactive({ + hvi <- hv_inputs() + hvi$envir <- r_data + do.call(homo_variance_test, hvi) +}) + +.summary_homo_variance_test <- reactive({ + if (hv_available() != "available") return(hv_available()) + summary(.homo_variance_test()) +}) + +.plot_homo_variance_test <- reactive({ + if (hv_available() != "available") return(hv_available()) + validate(need(input$hv_plots, i18n$t("Nothing to plot. Please select a plot type"))) + withProgress(message = i18n$t("Generating plots"), value = 1, + plot(.homo_variance_test(), plots = input$hv_plots, shiny = TRUE)) +}) + +## 10. Report +homo_variance_test_report <- function() { + if (is.empty(input$hv_var)) return(invisible()) + figs <- length(input$hv_plots) > 0 + outputs <- if (figs) c("summary", "plot") else "summary" + inp_out <- if (figs) list("", list(plots = input$hv_plots, custom = FALSE)) else list("", "") + update_report(inp_main = clean_args(hv_inputs(), hv_args), + fun_name = "homo_variance_test", + inp_out = inp_out, + outputs = outputs, + figs = figs, + fig.width = hv_plot_width(), + fig.height = hv_plot_height()) +} + +## 11. 下载 & 截图 +download_handler( + id = "dlp_homo_variance_test", + fun = download_handler_plot, + fn = function() paste0(input$dataset, "_homo_variance_test"), + type = "png", + caption = i18n$t("Save homogeneity of variance plot"), + plot = .plot_homo_variance_test, + width = hv_plot_width, + height = hv_plot_height +) + +observeEvent(input$homo_variance_test_report, { + r_info[["latest_screenshot"]] <- NULL + homo_variance_test_report() +}) + +observeEvent(input$homo_variance_test_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_homo_variance_test_screenshot") +}) + +observeEvent(input$modal_homo_variance_test_screenshot, { + homo_variance_test_report() + removeModal() +}) diff --git a/radiant.basics/inst/app/tools/analysis/normality_test_ui.R b/radiant.basics/inst/app/tools/analysis/normality_test_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..b22ad9aaab3acebc8c332718551cc55d3a859337 --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/normality_test_ui.R @@ -0,0 +1,181 @@ +############################################ +## Normality test - ui +############################################ + +## 1. 翻译标签 +nt_method <- c("shapiro", "ks", "ad") # 先给 3 个常用方法 +names(nt_method) <- c(i18n$t("Shapiro-Wilk"), + i18n$t("Kolmogorov-Smirnov"), + i18n$t("Anderson-Darling")) + +nt_plots <- c("qq", "hist", "pp", "density") +names(nt_plots) <- c(i18n$t("Q-Q plot"), + i18n$t("Histogram"), + i18n$t("P-P plot"), + i18n$t("Density")) + +## 2. 函数形参 +nt_args <- as.list(formals(normality_test)) + +## 3. 收集输入 +nt_inputs <- reactive({ + nt_args$data_filter <- if (input$show_filter) input$data_filter else "" + nt_args$dataset <- input$dataset + for (i in r_drop(names(nt_args))) { + nt_args[[i]] <- input[[paste0("nt_", i)]] + } + nt_args +}) + +## 4. 变量选择(仅 numeric) +output$ui_nt_var <- renderUI({ + isNum <- .get_class() %in% c("integer", "numeric", "ts") + vars <- c("None" = "", varnames()[isNum]) + selectInput( + inputId = "nt_var", label = i18n$t("Variable (select one):"), + choices = vars, selected = state_single("nt_var", vars), multiple = FALSE + ) +}) + +## 5. 主 UI +output$ui_normality_test <- renderUI({ + req(input$dataset) + tagList( + wellPanel( + conditionalPanel( + condition = "input.tabs_normality_test == 'Summary'", + uiOutput("ui_nt_var"), + selectInput( + inputId = "nt_method", label = i18n$t("Test method:"), + choices = nt_method, + selected = state_single("nt_method", nt_method, "shapiro"), + multiple = FALSE + ), + sliderInput( + "nt_conf_lev", i18n$t("Confidence level:"), + min = 0.85, max = 0.99, + value = state_init("nt_conf_lev", 0.95), step = 0.01 + ) + ), + conditionalPanel( + condition = "input.tabs_normality_test == 'Plot'", + selectizeInput( + inputId = "nt_plots", label = i18n$t("Select plots:"), + choices = nt_plots, + selected = state_multiple("nt_plots", nt_plots, "qq"), + multiple = TRUE, + options = list(placeholder = i18n$t("Select plots"), + plugins = list("remove_button", "drag_drop")) + ) + ) + ), + help_and_report( + modal_title = i18n$t("Normality test"), + fun_name = "normality_test", + help_file = inclMD(file.path(getOption("radiant.path.basics"), + "app/tools/help/normality_test.md")) + ) + ) +}) + +## 6. 画图尺寸(直接抄) +nt_plot <- reactive({ + list(plot_width = 650, + plot_height = 400 * max(length(input$nt_plots), 1)) +}) +nt_plot_width <- function() nt_plot()$plot_width +nt_plot_height <- function() nt_plot()$plot_height + + +## 7. 输出面板 +output$normality_test <- renderUI({ + register_print_output("summary_normality_test", ".summary_normality_test") + register_plot_output("plot_normality_test", ".plot_normality_test", + height_fun = "nt_plot_height") + + nt_output_panels <- tabsetPanel( + id = "tabs_normality_test", + tabPanel(title = i18n$t("Summary"), + value = "Summary", + verbatimTextOutput("summary_normality_test")), + tabPanel(title = i18n$t("Plot"), + value = "Plot", + download_link("dlp_normality_test"), + plotOutput("plot_normality_test", height = "100%")) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Normality"), + tool = i18n$t("Normality test"), + tool_ui = "ui_normality_test", + output_panels = nt_output_panels + ) +}) + +## 8. 可用性检查 +nt_available <- reactive({ + if (not_available(input$nt_var)) + return(i18n$t("This analysis requires a numeric variable. If none are\navailable please select another dataset.") %>% suggest_data("demand_uk")) + "available" +}) + +## 9. 计算核心 +.normality_test <- reactive({ + nti <- nt_inputs() + nti$envir <- r_data + do.call(normality_test, nti) +}) + +.summary_normality_test <- reactive({ + if (nt_available() != "available") return(nt_available()) + summary(.normality_test()) +}) + +.plot_normality_test <- reactive({ + if (nt_available() != "available") return(nt_available()) + validate(need(input$nt_plots, i18n$t("Nothing to plot. Please select a plot type"))) + withProgress(message = i18n$t("Generating plots"), value = 1, + plot(.normality_test(), plots = input$nt_plots, shiny = TRUE)) +}) + +## 10. Report +normality_test_report <- function() { + if (is.empty(input$nt_var)) return(invisible()) + figs <- length(input$nt_plots) > 0 + outputs <- if (figs) c("summary", "plot") else "summary" + inp_out <- if (figs) list("", list(plots = input$nt_plots, custom = FALSE)) else list("", "") + update_report(inp_main = clean_args(nt_inputs(), nt_args), + fun_name = "normality_test", + inp_out = inp_out, + outputs = outputs, + figs = figs, + fig.width = nt_plot_width(), + fig.height = nt_plot_height()) +} + +## 11. 下载 & 截图 +download_handler( + id = "dlp_normality_test", + fun = download_handler_plot, + fn = function() paste0(input$dataset, "_normality_test"), + type = "png", + caption = i18n$t("Save normality test plot"), + plot = .plot_normality_test, + width = nt_plot_width, + height = nt_plot_height +) + +observeEvent(input$normality_test_report, { + r_info[["latest_screenshot"]] <- NULL + normality_test_report() +}) + +observeEvent(input$normality_test_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_normality_test_screenshot") +}) + +observeEvent(input$modal_normality_test_screenshot, { + normality_test_report() + removeModal() +}) \ No newline at end of file diff --git a/radiant.basics/inst/app/tools/analysis/prob_calc_ui.R b/radiant.basics/inst/app/tools/analysis/prob_calc_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..a8422981f35e7af5e1154c412b7c9b539b36a341 --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/prob_calc_ui.R @@ -0,0 +1,572 @@ +pc_dist <- c("binom", "chisq", "disc", "expo", "fdist", "lnorm", "norm", "pois", "tdist", "unif") +names(pc_dist) <- c( + i18n$t("Binomial"), i18n$t("Chi-squared"), i18n$t("Discrete"), + i18n$t("Exponential"), i18n$t("F"), i18n$t("Log normal"), + i18n$t("Normal"), i18n$t("Poisson"), i18n$t("t"), i18n$t("Uniform") +) + +pc_type <- c("values", "probs") +names(pc_type) <- c(i18n$t("Values"), i18n$t("Probabilities")) + + +make_pc_values_input <- function(lb, lb_init = NA, ub, ub_init = 0) { + if(!is.empty(r_state[[lb]])) ub_init <- NA + if(!is.empty(r_state[[ub]])) lb_init <- NA + tags$table( + tags$td(numericInput(lb, i18n$t("Lower bound:"), value = state_init(lb, lb_init))), + tags$td(numericInput(ub, i18n$t("Upper bound:"), value = state_init(ub, ub_init))) + ) +} + +make_side_by_side <- function(a, b) { + tags$table( + tags$td(a, width="50%"), + tags$td(b, width="50%"), + width="100%" + ) +} + +make_pc_prob_input <- function(lb, lb_init = NA, ub, ub_init = 0.95) { + if(!is.empty(r_state[[lb]])) ub_init <- NA + if(!is.empty(r_state[[ub]])) lb_init <- NA + make_side_by_side( + numericInput( + lb, i18n$t("Lower bound:"), value = state_init(lb, lb_init), + min = 0, max = 1, step = .005 + ), + numericInput( + ub, i18n$t("Upper bound:"), value = state_init(ub, ub_init), + min = 0, max = 1, step = .005 + ) + ) +} + +output$ui_pc_pois <- renderUI({ + numericInput( + "pcp_lambda", i18n$t("Lambda:"), + value = state_init("pcp_lambda", 1), + min = 1 + ) +}) + +output$ui_pc_input_pois <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pcp_lb", lb_init = NA, "pcp_ub", ub_init = 3) + } else { + make_pc_prob_input("pcp_plb", lb_init = NA, "pcp_pub", ub_init = 0.95) + } +}) + +output$ui_pc_expo <- renderUI({ + numericInput( + "pce_rate", i18n$t("Rate:"), + value = state_init("pce_rate", 1), + min = 0 + ) +}) + +output$ui_pc_input_expo <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pce_lb", lb_init = NA, "pce_ub", ub_init = 2.996) + } else { + make_pc_prob_input("pce_plb", lb_init = NA, "pce_pub", ub_init = 0.95) + } +}) + +output$ui_pc_disc <- renderUI({ + tagList( + returnTextInput( + "pcd_v", i18n$t("Values:"), + value = state_init("pcd_v", "1 2 3 4 5 6") + ), + returnTextInput( + "pcd_p", i18n$t("Probabilities:"), + value = state_init("pcd_p", "1/6") + ) + ) +}) + +output$ui_pc_input_disc <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pcd_lb", lb_init = NA, "pcd_ub", ub_init = 3) + } else { + make_pc_prob_input("pcd_plb", lb_init = NA, "pcd_pub", ub_init = 0.95) + } +}) + +output$ui_pc_fdist <- renderUI({ + tagList( + numericInput( + "pcf_df1", i18n$t("Degrees of freedom 1:"), + value = state_init("pcf_df1", 10), + min = 1 + ), + numericInput( + "pcf_df2", i18n$t("Degrees of freedom 2:"), + value = state_init("pcf_df2", 10), + min = 5 + ) + ) +}) + +output$ui_pc_input_fdist <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pcf_lb", lb_init = NA, "pcf_ub", ub_init = 2.978) + } else { + make_pc_prob_input("pcf_plb", lb_init = NA, "pcf_pub", ub_init = 0.95) + } +}) + +output$ui_pc_chisq <- renderUI({ + numericInput( + "pcc_df", i18n$t("Degrees of freedom:"), + value = state_init("pcc_df", 1), + min = 1 + ) +}) + +output$ui_pc_input_chisq <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pcc_lb", lb_init = NA, "pcc_ub", ub_init = 3.841) + } else { + make_pc_prob_input("pcc_plb", lb_init = NA, "pcc_pub", ub_init = 0.95) + } +}) + +output$ui_pc_tdist <- renderUI({ + numericInput( + "pct_df", i18n$t("Degrees of freedom:"), + value = state_init("pct_df", 10), + min = 3 + ) +}) + +output$ui_pc_input_tdist <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pct_lb", lb_init = -Inf, "pct_ub", ub_init = 2.228) + } else { + make_pc_prob_input("pct_plb", lb_init = 0.025, "pct_pub", ub_init = 0.975) + } +}) + +output$ui_pc_norm <- renderUI({ + make_side_by_side( + numericInput( + "pc_mean", i18n$t("Mean:"), + value = state_init("pc_mean", 0) + ), + numericInput( + "pc_stdev", i18n$t("St. dev:"), + min = 0, + value = state_init("pc_stdev", 1) + ) + ) +}) + +output$ui_pc_input_norm <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pc_lb", lb_init = -Inf, "pc_ub", ub_init = 0) + } else { + make_pc_prob_input("pc_plb", lb_init = 0.025, "pc_pub", ub_init = 0.975) + } +}) + +output$ui_pc_lnorm <- renderUI({ + make_side_by_side( + numericInput( + "pcln_meanlog", i18n$t("Mean log:"), + value = state_init("pcln_meanlog", 0) + ), + numericInput( + "pcln_sdlog", i18n$t("St. dev log:"), + min = 0, + value = state_init("pcln_sdlog", 1) + ) + ) +}) + +output$ui_pc_input_lnorm <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pcln_lb", lb_init = 0, "pcln_ub", ub_init = 1) + } else { + make_pc_prob_input("pcln_plb", lb_init = 0.025, "pcln_pub", ub_init = 0.975) + } +}) + +output$ui_pc_binom <- renderUI({ + make_side_by_side( + numericInput( + "pcb_n", label = i18n$t("n:"), + value = state_init("pcb_n", 10), min = 0 + ), + numericInput( + "pcb_p", i18n$t("p:"), + min = 0, max = 1, step = .005, + value = state_init("pcb_p", .2) + ) + ) +}) + +output$ui_pc_input_binom <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pcb_lb", lb_init = NA, "pcb_ub", ub_init = 3) + } else { + make_pc_prob_input("pcb_plb", lb_init = NA, "pcb_pub", ub_init = 0.3) + } +}) + +output$ui_pc_unif <- renderUI({ + make_side_by_side( + numericInput( + "pcu_min", i18n$t("Min:"), + value = state_init("pcu_min", 0) + ), + numericInput( + "pcu_max", i18n$t("Max:"), + value = state_init("pcu_max", 1) + ) + ) +}) + +output$ui_pc_input_unif <- renderUI({ + if (input$pc_type == "values") { + make_pc_values_input("pcu_lb", lb_init = NA, "pcu_ub", ub_init = 0.3) + } else { + make_pc_prob_input("pcu_plb", lb_init = NA, "pcu_pub", ub_init = 0.3) + } +}) + +output$ui_prob_calc <- renderUI({ + tagList( + wellPanel( + selectInput( + "pc_dist", label = i18n$t("Distribution:"), + choices = pc_dist, + selected = state_init("pc_dist", "norm"), + multiple = FALSE + ), + conditionalPanel( + "input.pc_dist == 'norm'", + uiOutput("ui_pc_norm") + ), + conditionalPanel( + "input.pc_dist == 'lnorm'", + uiOutput("ui_pc_lnorm") + ), + conditionalPanel( + "input.pc_dist == 'binom'", + uiOutput("ui_pc_binom") + ), + conditionalPanel( + "input.pc_dist == 'unif'", + uiOutput("ui_pc_unif") + ), + conditionalPanel( + "input.pc_dist == 'tdist'", + uiOutput("ui_pc_tdist") + ), + conditionalPanel( + "input.pc_dist == 'fdist'", + uiOutput("ui_pc_fdist") + ), + conditionalPanel( + "input.pc_dist == 'chisq'", + uiOutput("ui_pc_chisq") + ), + conditionalPanel( + "input.pc_dist == 'disc'", + uiOutput("ui_pc_disc") + ), + conditionalPanel( + "input.pc_dist == 'expo'", + uiOutput("ui_pc_expo") + ), + conditionalPanel( + "input.pc_dist == 'pois'", + uiOutput("ui_pc_pois") + ) + ), + wellPanel( + radioButtons( + "pc_type", i18n$t("Input type:"), + choices = pc_type, + selected = state_init("pc_type", "values"), + inline = TRUE + ), + conditionalPanel( + "input.pc_dist == 'norm'", + uiOutput("ui_pc_input_norm") + ), + conditionalPanel( + "input.pc_dist == 'lnorm'", + uiOutput("ui_pc_input_lnorm") + ), + conditionalPanel( + "input.pc_dist == 'binom'", + uiOutput("ui_pc_input_binom") + ), + conditionalPanel( + "input.pc_dist == 'unif'", + uiOutput("ui_pc_input_unif") + ), + conditionalPanel( + "input.pc_dist == 'tdist'", + uiOutput("ui_pc_input_tdist") + ), + conditionalPanel( + "input.pc_dist == 'fdist'", + uiOutput("ui_pc_input_fdist") + ), + conditionalPanel( + "input.pc_dist == 'chisq'", + uiOutput("ui_pc_input_chisq") + ), + conditionalPanel( + "input.pc_dist == 'disc'", + uiOutput("ui_pc_input_disc") + ), + conditionalPanel( + "input.pc_dist == 'expo'", + uiOutput("ui_pc_input_expo") + ), + conditionalPanel( + "input.pc_dist == 'pois'", + uiOutput("ui_pc_input_pois") + ), + numericInput( + "pc_dec", i18n$t("Decimals:"), + value = state_init("pc_dec", 3), + min = 0 + ) + ), + help_and_report( + modal_title = i18n$t("Probability calculator"), + fun_name = "prob_calc", + help_file = inclMD(file.path(getOption("radiant.path.basics"), "app/tools/help/prob_calc.md")) + ) + ) +}) + +pc_plot_width <- function() + if (!is.null(input$viz_plot_width)) input$viz_plot_width else 650 + +pc_plot_height <- function() 400 + +pc_args <- reactive({ + pc_dist <- input$pc_dist + if (is.empty(pc_dist) || pc_dist == "norm") { + as.list(formals(prob_norm)) + } else if (pc_dist == "lnorm") { + as.list(formals(prob_lnorm)) + } else if (pc_dist == "binom") { + as.list(formals(prob_binom)) + } else if (pc_dist == "unif") { + as.list(formals(prob_unif)) + } else if (pc_dist == "tdist") { + as.list(formals(prob_tdist)) + } else if (pc_dist == "fdist") { + as.list(formals(prob_fdist)) + } else if (pc_dist == "chisq") { + as.list(formals(prob_chisq)) + } else if (pc_dist == "disc") { + as.list(formals(prob_disc)) + } else if (pc_dist == "expo") { + as.list(formals(prob_expo)) + } else if (pc_dist == "pois") { + as.list(formals(prob_pois)) + } +}) + +## list of function inputs selected by user +pc_inputs <- reactive({ + pc_dist <- input$pc_dist + if (is.empty(pc_dist) || pc_dist == "norm") { + pre <- "pc_" + } else if (pc_dist == "lnorm") { + pre <- "pcln_" + } else if (pc_dist == "binom") { + pre <- "pcb_" + } else if (pc_dist == "unif") { + pre <- "pcu_" + } else if (pc_dist == "tdist") { + pre <- "pct_" + } else if (pc_dist == "fdist") { + pre <- "pcf_" + } else if (pc_dist == "chisq") { + pre <- "pcc_" + } else if (pc_dist == "disc") { + pre <- "pcd_" + } else if (pc_dist == "expo") { + pre <- "pce_" + } else if (pc_dist == "pois") { + pre <- "pcp_" + } + + # loop needed because reactive values don't allow single bracket indexing + args <- pc_args() + for (i in names(args)) + args[[i]] <- input[[paste0(pre, i)]] + + validate( + need( + input$pc_dec, + i18n$t("Provide an integer value for the number of decimal places") + ) + ) + + args[["dec"]] <- input$pc_dec + args +}) + +## output is called from the main radiant ui.R +output$prob_calc <- renderUI({ + register_print_output("summary_prob_calc", ".summary_prob_calc") + register_plot_output( + "plot_prob_calc", ".plot_prob_calc", + height_fun = "pc_plot_height", + width_fun = "pc_plot_width" + ) + + ## two separate tabs + pc_output_panels <- tagList( + tabPanel(i18n$t("Summary"), value = "Summary", verbatimTextOutput("summary_prob_calc")), + tabPanel( + i18n$t("Plot"), value = "Plot", + download_link("dlp_prob_calc"), + plotOutput("plot_prob_calc", width = "100%", height = "100%") + ) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Probability"), + tool = i18n$t("Probability calculator"), + data = NULL, + tool_ui = "ui_prob_calc", + output_panels = pc_output_panels + ) +}) + +pc_available <- reactive({ + if (is.empty(input$pc_dist) || is.empty(input$pc_type)) { + "" + } else { + a <- "available" + if (input$pc_dist == "norm") { + if (is_not(input$pc_mean) || is_not(input$pc_stdev) || input$pc_stdev <= 0) { + a <- i18n$t("Please provide a mean and standard deviation (> 0)") + } + } else if (input$pc_dist == "lnorm") { + if (is_not(input$pcln_meanlog) || is_not(input$pcln_sdlog) || input$pcln_sdlog <= 0) { + a <- i18n$t("Please provide a mean and standard deviation (> 0)") + } + } else if (input$pc_dist == "binom") { + if (is_not(input$pcb_n) || input$pcb_n < 0 || is_not(input$pcb_p) || input$pcb_p < 0) { + a <- i18n$t("Please provide a value for n (number of trials) and p (probability of success)") + } + } else if (input$pc_dist == "unif") { + if (is_not(input$pcu_min) || is_not(input$pcu_max)) { + a <- i18n$t("Please provide a minimum and a maximum value") + } + } else if (input$pc_dist == "tdist") { + if (is_not(input$pct_df)) { + a <- i18n$t("Please provide a value for the degrees of freedom (> 0)") + } + } else if (input$pc_dist == "fdist") { + if (is_not(input$pcf_df1) || is_not(input$pcf_df2) || input$pcf_df1 < 1 || input$pcf_df2 < 5) { + a <- i18n$t("Please provide a value for Degrees of freedom 1 (> 0)\nand for Degrees of freedom 2 (> 4)") + } + } else if (input$pc_dist == "chisq") { + if (is_not(input$pcc_df)) { + a <- i18n$t("Please provide a value for the degrees of freedom (> 0)") + } + } else if (input$pc_dist == "disc") { + if (is.empty(input$pcd_v) || is.empty(input$pcd_p)) { + a <- i18n$t("Please provide a set of values and probabilities.\nSeparate numbers using spaces (e.g., 1/2 1/2)") + } + } else if (input$pc_dist == "expo") { + if (is_not(input$pce_rate) || input$pce_rate <= 0) { + a <- i18n$t("Please provide a value for the rate (> 0)") + } + } else if (input$pc_dist == "pois") { + if (is_not(input$pcp_lambda) || input$pcp_lambda <= 0) { + a <- i18n$t("Please provide a value for lambda (> 0)") + } + } else { + a <- "" + } + a + } +}) + +.prob_calc <- reactive({ + validate( + need(pc_available() == "available", pc_available()) + ) + do.call(get(paste0("prob_", input$pc_dist)), pc_inputs()) +}) + +.summary_prob_calc <- reactive({ + type <- if (is.empty(input$pc_type)) "values" else input$pc_type + summary(.prob_calc(), type = type) +}) + +.plot_prob_calc <- reactive({ + req(pc_available() == "available") + type <- if (is.empty(input$pc_type)) "values" else input$pc_type + plot(.prob_calc(), type = type) +}) + +prob_calc_report <- function() { + req(input$pc_dist) + type <- input$pc_type + inp <- pc_inputs() + if (!is.null(type) && type == "probs") { + inp_out <- list(type = type) %>% list(., .) + inp[["ub"]] <- inp[["lb"]] <- NA + } else { + inp_out <- list("", "") + inp[["pub"]] <- inp[["plb"]] <- NA + } + + if (input$pc_dist == "disc") { + inp$v <- radiant.data::make_vec(inp$v) + inp$p <- radiant.data::make_vec(inp$p) + } + + outputs <- c("summary", "plot") + update_report( + inp_main = clean_args(inp, pc_args()), + fun_name = paste0("prob_", input$pc_dist), + inp_out = inp_out, + outputs = outputs, + figs = TRUE, + fig.width = pc_plot_width(), + fig.height = pc_plot_height() + ) +} + +download_handler( + id = "dlp_prob_calc", + fun = download_handler_plot, + fn = function() paste0(input$pc_dist, "_prob_calc"), + type = "png", + caption = i18n$t("Save probability calculator plot"), + plot = .plot_prob_calc, + width = pc_plot_width, + height = pc_plot_height +) + +observeEvent(input$prob_calc_report, { + r_info[["latest_screenshot"]] <- NULL + prob_calc_report() +}) + +observeEvent(input$prob_calc_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_prob_calc_screenshot") +}) + +observeEvent(input$modal_prob_calc_screenshot, { + prob_calc_report() + removeModal() ## remove shiny modal after save +}) diff --git a/radiant.basics/inst/app/tools/analysis/single_mean_ui.R b/radiant.basics/inst/app/tools/analysis/single_mean_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..7dd125927abaa2b746519385a0b91b594bcea09e --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/single_mean_ui.R @@ -0,0 +1,201 @@ +############################### +## Single mean - ui +############################### + +## alternative hypothesis options +sm_alt <- c("two.sided", "less", "greater") +names(sm_alt) <- c(i18n$t("Two sided"), i18n$t("Less than"), i18n$t("Greater than")) + +sm_plots <- c("hist", "simulate") +names(sm_plots) <- c(i18n$t("Histogram"), i18n$t("Simulate")) + +## list of function arguments +sm_args <- as.list(formals(single_mean)) + +## list of function inputs selected by user +sm_inputs <- reactive({ + ## loop needed because reactive values don't allow single bracket indexing + sm_args$data_filter <- if (input$show_filter) input$data_filter else "" + sm_args$dataset <- input$dataset + for (i in r_drop(names(sm_args))) { + sm_args[[i]] <- input[[paste0("sm_", i)]] + } + sm_args +}) + +output$ui_sm_var <- renderUI({ + isNum <- .get_class() %in% c("integer", "numeric", "ts") + vars <- c("None" = "", varnames()[isNum]) + selectInput( + inputId = "sm_var", label = i18n$t("Variable (select one):"), + choices = vars, selected = state_single("sm_var", vars), multiple = FALSE + ) +}) + +output$ui_single_mean <- renderUI({ + req(input$dataset) + tagList( + wellPanel( + conditionalPanel( + condition = "input.tabs_single_mean == 'Summary'", + uiOutput("ui_sm_var"), + selectInput( + inputId = "sm_alternative", label = i18n$t("Alternative hypothesis:"), + choices = sm_alt, + selected = state_single("sm_alternative", sm_alt, sm_args$alternative), + multiple = FALSE + ), + sliderInput( + "sm_conf_lev", i18n$t("Confidence level:"), + min = 0.85, max = 0.99, + value = state_init("sm_conf_lev", sm_args$conf_lev), step = 0.01 + ), + numericInput( + "sm_comp_value", i18n$t("Comparison value:"), + state_init("sm_comp_value", sm_args$comp_value) + ) + ), + conditionalPanel( + condition = "input.tabs_single_mean == 'Plot'", + selectizeInput( + inputId = "sm_plots", label = i18n$t("Select plots:"), + choices = sm_plots, + selected = state_multiple("sm_plots", sm_plots, "hist"), + multiple = TRUE, + options = list(placeholder = i18n$t("Select plots"), plugins = list("remove_button", "drag_drop")) + ) + ) + ), + help_and_report( + modal_title = i18n$t("Single mean"), + fun_name = "single_mean", + help_file = inclMD(file.path(getOption("radiant.path.basics"), "app/tools/help/single_mean.md")) + ) + ) +}) + +sm_plot <- reactive({ + list(plot_width = 650, plot_height = 400 * max(length(input$sm_plots), 1)) +}) + +sm_plot_width <- function() { + sm_plot() %>% + { + if (is.list(.)) .$plot_width else 650 + } +} + +sm_plot_height <- function() { + sm_plot() %>% + { + if (is.list(.)) .$plot_height else 400 + } +} + +## output is called from the main radiant ui.R +output$single_mean <- renderUI({ + register_print_output("summary_single_mean", ".summary_single_mean") + register_plot_output( + "plot_single_mean", ".plot_single_mean", + height_fun = "sm_plot_height" + ) + + ## two separate tabs + sm_output_panels <- tabsetPanel( + id = "tabs_single_mean", + tabPanel(i18n$t("Summary"), value = "Summary", verbatimTextOutput("summary_single_mean")), + tabPanel( + i18n$t("Plot"), value = "Plot", + download_link("dlp_single_mean"), + plotOutput("plot_single_mean", height = "100%") + ) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Means"), + tool = i18n$t("Single mean"), + tool_ui = "ui_single_mean", + output_panels = sm_output_panels + ) +}) + +sm_available <- reactive({ + if (not_available(input$sm_var)) { + i18n$t("This analysis requires a variable of type numeric or interval. If none are\navailable please select another dataset.\n\n") %>% suggest_data("demand_uk") + } else if (is.na(input$sm_comp_value)) { + i18n$t("Please choose a comparison value") + } else { + "available" + } +}) + +.single_mean <- reactive({ + smi <- sm_inputs() + smi$envir <- r_data + do.call(single_mean, smi) +}) + +.summary_single_mean <- reactive({ + if (sm_available() != "available") { + return(sm_available()) + } + summary(.single_mean()) +}) + +.plot_single_mean <- reactive({ + if (sm_available() != "available") { + return(sm_available()) + } + validate(need(input$sm_plots, i18n$t("Nothing to plot. Please select a plot type"))) + withProgress(message = i18n$t("Generating plots"), value = 1, { + plot(.single_mean(), plots = input$sm_plots, shiny = TRUE) + }) +}) + +single_mean_report <- function() { + if (is.empty(input$sm_var)) { + return(invisible()) + } + if (length(input$sm_plots) == 0) { + figs <- FALSE + outputs <- c("summary") + inp_out <- list("", "") + } else { + outputs <- c("summary", "plot") + inp_out <- list("", list(plots = input$sm_plots, custom = FALSE)) + figs <- TRUE + } + update_report( + inp_main = clean_args(sm_inputs(), sm_args), + fun_name = "single_mean", inp_out = inp_out, + outputs = outputs, figs = figs, + fig.width = sm_plot_width(), + fig.height = sm_plot_height() + ) +} + +download_handler( + id = "dlp_single_mean", + fun = download_handler_plot, + fn = function() paste0(input$dataset, "_single_mean"), + type = "png", + caption = i18n$t("Save single mean plot"), + plot = .plot_single_mean, + width = sm_plot_width, + height = sm_plot_height +) + +observeEvent(input$single_mean_report, { + r_info[["latest_screenshot"]] <- NULL + single_mean_report() +}) + +observeEvent(input$single_mean_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_single_mean_screenshot") +}) + +observeEvent(input$modal_single_mean_screenshot, { + single_mean_report() + removeModal() ## remove shiny modal after save +}) diff --git a/radiant.basics/inst/app/tools/analysis/single_prop_ui.R b/radiant.basics/inst/app/tools/analysis/single_prop_ui.R new file mode 100644 index 0000000000000000000000000000000000000000..df99c83cfff6c52164eb0a9d2fae26478a0f3c80 --- /dev/null +++ b/radiant.basics/inst/app/tools/analysis/single_prop_ui.R @@ -0,0 +1,228 @@ +############################### +# Single proportion - ui +############################### + +## alternative hypothesis options +sp_alt <- list("two.sided", "less", "greater") +names(sp_alt) <- c( + i18n$t("Two sided"), + i18n$t("Less than"), + i18n$t("Greater than") +) +sp_plots <- c("bar", "simulate") +names(sp_plots) <- c(i18n$t("Bar"), i18n$t("Simulate")) + +## list of function arguments +sp_args <- as.list(formals(single_prop)) + +## list of function inputs selected by user +sp_inputs <- reactive({ + ## loop needed because reactive values don't allow single bracket indexing + sp_args$data_filter <- if (input$show_filter) input$data_filter else "" + sp_args$dataset <- input$dataset + for (i in r_drop(names(sp_args))) { + sp_args[[i]] <- input[[paste0("sp_", i)]] + } + sp_args +}) + +output$ui_sp_var <- renderUI({ + vars <- c("None" = "", groupable_vars()) + selectInput( + inputId = "sp_var", label = i18n$t("Variable (select one):"), + choices = vars, + selected = state_single("sp_var", vars), + multiple = FALSE + ) +}) + +output$up_sp_lev <- renderUI({ + req(available(input$sp_var)) + levs <- .get_data()[[input$sp_var]] %>% + as.factor() %>% + levels() + + selectInput( + "sp_lev", i18n$t("Choose level:"), + choices = levs, + selected = state_single("sp_lev", levs), + multiple = FALSE + ) +}) + +output$ui_single_prop <- renderUI({ + req(input$dataset) + tagList( + wellPanel( + conditionalPanel( + condition = "input.tabs_single_prop == 'Summary'", + uiOutput("ui_sp_var"), + uiOutput("up_sp_lev"), + selectInput( + "sp_alternative", i18n$t("Alternative hypothesis:"), + choices = sp_alt, + selected = state_single("sp_alternative", sp_alt, sp_args$alternative), + multiple = FALSE + ), + sliderInput( + "sp_conf_lev", i18n$t("Confidence level:"), + min = 0.85, max = 0.99, step = 0.01, + value = state_init("sp_conf_lev", sp_args$conf_lev) + ), + numericInput( + "sp_comp_value", i18n$t("Comparison value:"), + value = state_init("sp_comp_value", sp_args$comp_value), + min = 0.01, max = 0.99, step = 0.01 + ), + # radioButtons("sp_type", label = "Test:", c("Binomial" = "binom", "Chi-square" = "chisq"), + radioButtons( + inputId = "sp_test", + label = i18n$t("Test type:"), + choices = { + opts <- c("binom", "z") + names(opts) <- c(i18n$t("Binomial exact"), i18n$t("Z-test")) + opts + }, + selected = state_init("sp_test", "binom"), inline = TRUE + ) + ), + conditionalPanel( + condition = "input.tabs_single_prop == 'Plot'", + selectizeInput( + "sp_plots", i18n$t("Select plots:"), + choices = sp_plots, + selected = state_multiple("sp_plots", sp_plots, "bar"), + multiple = TRUE, + options = list(placeholder = i18n$t("Select plots"), plugins = list("remove_button", "drag_drop")) + ) + ) + ), + help_and_report( + modal_title = i18n$t("Single proportion"), + fun_name = "single_prop", + help_file = inclMD(file.path(getOption("radiant.path.basics"), "app/tools/help/single_prop.md")) + ) + ) +}) + +sp_plot <- reactive({ + list(plot_width = 650, plot_height = 400 * max(length(input$sp_plots), 1)) +}) + +sp_plot_width <- function() { + sp_plot() %>% + (function(x) if (is.list(x)) x$plot_width else 650) +} + +sp_plot_height <- function() { + sp_plot() %>% + (function(x) if (is.list(x)) x$plot_height else 400) +} + +## output is called from the main radiant ui.R +output$single_prop <- renderUI({ + register_print_output("summary_single_prop", ".summary_single_prop") + register_plot_output( + "plot_single_prop", ".plot_single_prop", + height_fun = "sp_plot_height" + ) + + ## two separate tabs + sp_output_panels <- tabsetPanel( + id = "tabs_single_prop", + tabPanel(i18n$t("Summary"), value = "Summary", verbatimTextOutput("summary_single_prop")), + tabPanel( + i18n$t("Plot"), value = "Plot", + download_link("dlp_single_prop"), + plotOutput("plot_single_prop", height = "100%") + ) + ) + + stat_tab_panel( + menu = i18n$t("Basics > Proportions"), + tool = i18n$t("Single proportion"), + tool_ui = "ui_single_prop", + output_panels = sp_output_panels + ) +}) + +sp_available <- reactive({ + if (not_available(input$sp_var)) { + i18n$t("This analysis requires a categorical variable. In none are available\nplease select another dataset.\n\n") %>% suggest_data("consider") + } else if (input$sp_comp_value %>% (function(x) is.na(x) | x > 1 | x <= 0)) { + i18n$t("Please choose a comparison value between 0 and 1") + } else { + "available" + } +}) + +.single_prop <- reactive({ + spi <- sp_inputs() + spi$envir <- r_data + do.call(single_prop, spi) +}) + +.summary_single_prop <- reactive({ + if (sp_available() != "available") { + return(sp_available()) + } + summary(.single_prop()) +}) + +.plot_single_prop <- reactive({ + if (sp_available() != "available") { + return(sp_available()) + } + validate(need(input$sp_plots, i18n$t("Nothing to plot. Please select a plot type"))) + withProgress(message = i18n$t("Generating plots"), value = 1, { + plot(.single_prop(), plots = input$sp_plots, shiny = TRUE) + }) +}) + +single_prop_report <- function() { + if (is.empty(input$sp_var)) { + return(invisible()) + } + if (length(input$sp_plots) == 0) { + figs <- FALSE + outputs <- c("summary") + inp_out <- list("", "") + } else { + outputs <- c("summary", "plot") + inp_out <- list("", list(plots = input$sp_plots, custom = FALSE)) + figs <- TRUE + } + update_report( + inp_main = clean_args(sp_inputs(), sp_args), + fun_name = "single_prop", inp_out = inp_out, + outputs = outputs, figs = figs, + fig.width = sp_plot_width(), + fig.height = sp_plot_height() + ) +} + +download_handler( + id = "dlp_single_prop", + fun = download_handler_plot, + fn = function() paste0(input$dataset, "_single_prop"), + type = "png", + caption = i18n$t("Save single proportion plot"), + plot = .plot_single_prop, + width = sp_plot_width, + height = sp_plot_height +) + +observeEvent(input$single_prop_report, { + r_info[["latest_screenshot"]] <- NULL + single_prop_report() +}) + +observeEvent(input$single_prop_screenshot, { + r_info[["latest_screenshot"]] <- NULL + radiant_screenshot_modal("modal_single_prop_screenshot") +}) + +observeEvent(input$modal_single_prop_screenshot, { + single_prop_report() + removeModal() ## remove shiny modal after save +}) diff --git a/radiant.basics/inst/app/tools/help/clt.md b/radiant.basics/inst/app/tools/help/clt.md new file mode 100644 index 0000000000000000000000000000000000000000..a446ed5ab9cfb947621ca30f497c234ea639a8de --- /dev/null +++ b/radiant.basics/inst/app/tools/help/clt.md @@ -0,0 +1,19 @@ +> 用随机抽样说明中心极限定理 + +### 什么是中心极限定理? + +“在概率论中,中心极限定理(CLT)指出,在特定条件下,大量独立随机变量(每个变量都有明确的期望值和方差)的算术平均值将近似服从正态分布,而与变量的潜在分布无关。也就是说,假设获取一个包含大量观测值的样本,每个观测值都是随机生成的,且不依赖于其他观测值的值,然后计算观测值的算术平均值。如果多次执行此过程,中心极限定理表明,计算得到的平均值将服从正态分布(通常称为‘钟形曲线’)。” + +来源:维基百科 + +## 抽样 + +要生成样本,请从 “分布(Distribution)” 下拉菜单中选择一种分布,并接受(或更改)默认值。然后点击 “抽样(Sample)” 按钮或按`CTRL-enter`(Mac 上为`CMD-enter`)运行模拟并显示模拟数据的图表。 + +### Khan 讲解中心极限定理 + + + +### R 函数 + +有关 Radiant 中用于概率计算的相关 R 函数概述,请参见*基础 > 概率* 。 \ No newline at end of file diff --git a/radiant.basics/inst/app/tools/help/compare_means.md b/radiant.basics/inst/app/tools/help/compare_means.md new file mode 100644 index 0000000000000000000000000000000000000000..be4a420988864803fefe200446bc56adb8b9665e --- /dev/null +++ b/radiant.basics/inst/app/tools/help/compare_means.md @@ -0,0 +1,120 @@ +> 比较数据中两个或多个变量或组的均值 + +均值比较 t 检验用于比较一个组中某个变量的均值与一个或多个其他组中同一变量的均值。总体中组间差异的原假设设为零。我们使用样本数据检验这一假设。 + +我们可以执行单侧检验(即`小于`或`大于`)或双侧检验(见 “备择假设(Alternative hypothesis)” 下拉菜单)。单侧检验用于评估现有数据是否提供证据表明组间样本均值差异小于(或大于)零。 + +### 示例:教授薪资 + +我们获取了美国某学院助理教授、副教授和教授的 9 个月学术薪资数据(2008-09 学年)。这些数据是学院行政部门为监测男女教师薪资差异而持续收集的一部分。数据包含 397 个观测值和以下 6 个变量: + +- `rank` = 因子,水平为 AsstProf(助理教授)、AssocProf(副教授)、Prof(教授) +- `discipline` = 因子,水平为 A(“理论型” 院系)或 B(“应用型” 院系) +- `yrs.since.phd` = 获得博士学位后的年数 +- `yrs.service` = 任职年数 +- `sex` = 因子,水平为 Female(女性)和 Male(男性) +- `salary` = 9 个月薪资(美元) + +这些数据来自 CAR 包,与以下书籍相关:Fox J. 和 Weisberg, S. (2011)《应用回归的 R 伴侣(第二版)》,Sage 出版社。 + +假设我们要检验职级较低的教授是否比职级较高的教授薪资更低。为检验这一假设,我们首先选择教授`rank`,并选择`salary`作为要在不同职级间比较的数值变量。在 “选择组合(Choose combinations)” 框中选择所有可用条目,对三个职级进行两两比较。注意,移除所有条目会自动选择所有组合。我们关注单侧假设(即`小于`)。 + +

+Pairwise mean comparisons (t-test) +Data : salary +Variables : rank, salary +Samples : independent +Confidence: 0.95 +Adjustment: None + + rank mean n n_missing sd se me + AsstProf 80,775.985 67 0 8,174.113 998.627 1,993.823 + AssocProf 93,876.438 64 0 13,831.700 1,728.962 3,455.056 + Prof 126,772.109 266 0 27,718.675 1,699.541 3,346.322 + + Null hyp. Alt. hyp. diff p.value se t.value df 0% 95% + AsstProf = AssocProf AsstProf < AssocProf -13100.45 < .001 1996.639 -6.561 101.286 -Inf -9785.958 *** + AsstProf = Prof AsstProf < Prof -45996.12 < .001 1971.217 -23.334 324.340 -Inf -42744.474 *** + AssocProf = Prof AssocProf < Prof -32895.67 < .001 2424.407 -13.569 199.325 -Inf -28889.256 *** + +Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ++ +* `se`是标准误(即`diff`抽样分布的标准差) +* `t.value`是与`diff`相关的 t 统计量,可与 t 分布比较(即`diff` / `se`) +* `df`是统计检验的自由度。注意,自由度使用 Welch 近似法计算 +* `0% 95%`显示样本均值差异的 95% 置信区间。这些数值提供了真实总体差异可能落入的范围 + +### 检验方法 + +我们可以使用三种方法评估原假设。我们选择显著性水平为 0.05。1 当然,每种方法会得出相同结论。 + +#### p 值 + +由于每个 p 值都**小于**显著性水平,我们拒绝每个评估的教授职级对的原假设。数据表明,副教授薪资高于助理教授,教授薪资高于助理教授和副教授。注意,“***” 用作显著性指标。 + +#### 置信区间 + +由于任何置信区间都**不**包含零,我们拒绝每个评估的职级组合的原假设。由于我们的备择假设是`小于`,置信区间实际上是总体薪资差异的 95% 置信上限(即 - 9785.958、-42744.474 和 - 28889.256)。 + +#### t 值 + +由于计算的 t 值(-6.561、-23.334 和 - 13.569)**小于**相应的临界 t 值,我们拒绝每个评估的职级组合的原假设。可通过 “基础(Basics)” 菜单中的概率计算器获取临界 t 值。以助理教授与副教授的检验为例,我们发现对于自由度为 101.286 的 t 分布(见`df`),临界 t 值为 1.66。由于备择假设是`小于`,我们选择 0.05 作为下侧概率界。 + +


usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+均值比较假设检验
+
+- 本视频展示如何进行均值比较假设检验
+- 主题列表:
+ - 按组计算汇总统计量
+ - 在 Radiant 中设置均值比较的假设检验
+ - 使用 p 值和置信区间评估假设检验
diff --git a/radiant.basics/inst/app/tools/help/compare_props.md b/radiant.basics/inst/app/tools/help/compare_props.md
new file mode 100644
index 0000000000000000000000000000000000000000..d309afe4b2fba570b40483eb72eff7a4e3c3d9f2
--- /dev/null
+++ b/radiant.basics/inst/app/tools/help/compare_props.md
@@ -0,0 +1,119 @@
+> 比较数据中两个或多个组的比例
+
+比例比较检验用于评估某些事件、行为、意图等的发生频率在不同组间是否存在差异。总体中组间比例差异的原假设设为零。我们使用样本数据检验这一假设。
+
+我们可以执行单侧检验(即`小于`或`大于`)或双侧检验(见 “备择假设(Alternative hypothesis)” 下拉菜单)。单侧检验适用于评估样本数据是否表明,例如,某一无线运营商的掉话比例比其他运营商更高(或更低)。
+
+### 示例
+
+我们将使用泰坦尼克号乘客生存状态数据集的一个样本。泰坦尼克号乘客数据的主要来源是《泰坦尼克号百科全书》。原始来源之一是 Eaton & Haas(1994)的《泰坦尼克号:胜利与悲剧》(Patrick Stephens Ltd 出版),其中包含由多位研究者整理、经 Michael A. Findlay 编辑的乘客名单。我们关注数据中的两个变量:
+
+- `survived` = 因子,水平为`Yes`(是)和`No`(否)
+- `pclass` = 乘客等级(1 等、2 等、3 等),作为社会经济地位(SES)的替代指标:1 等≈上层;2 等≈中层;3 等≈下层
+
+假设我们要检验泰坦尼克号沉没事件中,不同乘客等级的生存比例是否存在差异。为检验这一假设,我们选择`pclass`作为分组变量,并计算`survived`中`yes`的比例(见 “选择水平(Choose level)”)(见 “变量(选择一个)(Variable (select one))”)。
+
+在 “选择组合(Choose combinations)” 框中选择所有可用条目,对三个乘客等级进行两两比较。注意,移除所有条目会自动选择所有组合。除非我们对效应方向有明确假设,否则应使用双侧检验(即`two.sided`)。我们的第一个备择假设是 “1 等舱乘客的生存比例与 2 等舱乘客不同”。
+
+
+Pairwise proportion comparisons +Data : titanic +Variables : pclass, survived +Level : Yes in survived +Confidence: 0.95 +Adjustment: None + + pclass Yes No p n n_missing sd se me + 1st 179 103 0.635 282 0 8.086 0.029 0.056 + 2nd 115 146 0.441 261 0 8.021 0.031 0.060 + 3rd 131 369 0.262 500 0 9.832 0.020 0.039 + + Null hyp. Alt. hyp. diff p.value chisq.value df 2.5% 97.5% + 1st = 2nd 1st not equal to 2nd 0.194 < .001 20.576 1 0.112 0.277 *** + 1st = 3rd 1st not equal to 3rd 0.373 < .001 104.704 1 0.305 0.441 *** + 2nd = 3rd 2nd not equal to 3rd 0.179 < .001 25.008 1 0.107 0.250 *** + +Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ++ +* `chisq.value`是与`diff`相关的卡方统计量,可与卡方分布比较。关于该指标的计算方法,详见 “基础> 表格 > 交叉表” 的帮助文件。每组组合都会计算等效的 2×2 交叉表。 +* `df`是每个统计检验的自由度(1)。 +* `2.5% 97.5%`显示样本比例差异的 95% 置信区间。这些数值提供了真实总体差异可能落入的范围。 + +### 检验方法 + +我们可以使用三种方法评估原假设。我们选择显著性水平为 0.05。1 当然,每种方法会得出相同结论。 + +#### p 值 + +由于每个两两比较的 p 值都**小于**显著性水平,基于可用样本数据,我们可以拒绝比例相等的原假设。结果表明,1 等舱乘客比 2 等舱和 3 等舱乘客更可能在沉没事件中幸存;同样,2 等舱乘客比 3 等舱乘客更可能幸存。 + +#### 置信区间 + +由于任何置信区间都**不**包含零,我们拒绝每个评估的乘客等级组合的原假设。 + +#### 卡方值 + +由于计算的卡方值(20.576、104.704 和 25.008)**大于**相应的临界卡方值,我们拒绝每个评估的乘客等级组合的原假设。可通过 “基础(Basics)” 菜单中的概率计算器获取临界卡方值。以 1 等舱与 2 等舱乘客的检验为例,我们发现对于自由度为 1(见`df`)、置信水平为 0.95 的卡方分布,临界卡方值为 3.841。 + +


usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+比例比较假设检验
+
+- 本视频展示如何进行比例比较假设检验
+- 主题列表:
+ - 在 Radiant 中设置比例比较的假设检验
+ - 使用 p 值和置信区间评估假设检验
diff --git a/radiant.basics/inst/app/tools/help/correlation.md b/radiant.basics/inst/app/tools/help/correlation.md
new file mode 100644
index 0000000000000000000000000000000000000000..9520c3e1f5eac77c95ec093b357bbc7b76de4d64
--- /dev/null
+++ b/radiant.basics/inst/app/tools/help/correlation.md
@@ -0,0 +1,54 @@
+> 数据中变量的相关性如何?
+
+创建所选变量的相关矩阵。为每个变量对提供相关性和 p 值。要仅显示高于特定(绝对)水平的相关性,使用相关性截断框。
+
+注意:相关性可基于`numeric`、`integer`、`date`和`factor`类型的变量计算。当纳入因子型变量时,应勾选 “调整因子型变量(Adjust for {factor} variables)” 框。进行调整后估计相关性时,因子型变量将被视为(有序)分类变量,其他所有变量将被视为连续变量。
+
+






usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+交叉表假设检验
+
+- 本视频演示如何通过交叉表假设检验研究两个分类变量之间的关联
+- 主题列表:
+ - 在 Radiant 中设置交叉表的假设检验
+ - 解释观察频数表、期望频数表和卡方贡献表的构建方式
+ - 使用 p 值和临界值评估假设检验
diff --git a/radiant.basics/inst/app/tools/help/figures/compare_means_plot.png b/radiant.basics/inst/app/tools/help/figures/compare_means_plot.png
new file mode 100644
index 0000000000000000000000000000000000000000..78038008b60cd31b4eddaf6a4af9ab3d434b6563
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/compare_means_plot.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/compare_means_prob_calc.png b/radiant.basics/inst/app/tools/help/figures/compare_means_prob_calc.png
new file mode 100644
index 0000000000000000000000000000000000000000..23277042ca69f06afbdcbda9e2c51f91e531a260
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/compare_means_prob_calc.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/compare_means_summary.png b/radiant.basics/inst/app/tools/help/figures/compare_means_summary.png
new file mode 100644
index 0000000000000000000000000000000000000000..1248700dc6ad0b6ea33057081b5d772d199bf50c
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/compare_means_summary.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/compare_means_summary_additional.png b/radiant.basics/inst/app/tools/help/figures/compare_means_summary_additional.png
new file mode 100644
index 0000000000000000000000000000000000000000..0bf7e4d4314f57f32aab2c12e9f502afc1a4f85a
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/compare_means_summary_additional.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/compare_props_plot.png b/radiant.basics/inst/app/tools/help/figures/compare_props_plot.png
new file mode 100644
index 0000000000000000000000000000000000000000..8cf24791321ea0170418447208ac1b5ce54d7331
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/compare_props_plot.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/compare_props_prob_calc.png b/radiant.basics/inst/app/tools/help/figures/compare_props_prob_calc.png
new file mode 100644
index 0000000000000000000000000000000000000000..e7e373bd109429e67e2e364e76bd26e0bfd25f36
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/compare_props_prob_calc.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/compare_props_summary.png b/radiant.basics/inst/app/tools/help/figures/compare_props_summary.png
new file mode 100644
index 0000000000000000000000000000000000000000..89f8e70eca8a9c6e5483933317ef04520a8e2649
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/compare_props_summary.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/compare_props_summary_additional.png b/radiant.basics/inst/app/tools/help/figures/compare_props_summary_additional.png
new file mode 100644
index 0000000000000000000000000000000000000000..6e7aa40e94065dfaa1ea55e888b33a1c509ded3d
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/compare_props_summary_additional.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/correlation_plot.png b/radiant.basics/inst/app/tools/help/figures/correlation_plot.png
new file mode 100644
index 0000000000000000000000000000000000000000..b808e353362c2210818ecc624b5c31e81e298d8d
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/correlation_plot.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/correlation_store.png b/radiant.basics/inst/app/tools/help/figures/correlation_store.png
new file mode 100644
index 0000000000000000000000000000000000000000..596b87fc6a50c1095c4d586797c0dbf6688437f6
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/correlation_store.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/correlation_summary.png b/radiant.basics/inst/app/tools/help/figures/correlation_summary.png
new file mode 100644
index 0000000000000000000000000000000000000000..7bbbadb53eac4eeb5061cd6a37f160bb0c123c3a
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/correlation_summary.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/cross_tabs_chi_critical.png b/radiant.basics/inst/app/tools/help/figures/cross_tabs_chi_critical.png
new file mode 100644
index 0000000000000000000000000000000000000000..f3fda78bc06a26b9e1225153fd530b2e6045a02e
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/cross_tabs_chi_critical.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/cross_tabs_chi_pvalue.png b/radiant.basics/inst/app/tools/help/figures/cross_tabs_chi_pvalue.png
new file mode 100644
index 0000000000000000000000000000000000000000..adee22a4a080b4c642e6395e53b14d359a5a5de0
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/cross_tabs_chi_pvalue.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/cross_tabs_plot.png b/radiant.basics/inst/app/tools/help/figures/cross_tabs_plot.png
new file mode 100644
index 0000000000000000000000000000000000000000..35fdafd244096b2a08a2921803dd92d732062e3b
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/cross_tabs_plot.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/cross_tabs_summary.png b/radiant.basics/inst/app/tools/help/figures/cross_tabs_summary.png
new file mode 100644
index 0000000000000000000000000000000000000000..8d62a388d5eff23eb5bf3c1ac628d98de25c45e4
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/cross_tabs_summary.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/goodness_chi_pvalue.png b/radiant.basics/inst/app/tools/help/figures/goodness_chi_pvalue.png
new file mode 100644
index 0000000000000000000000000000000000000000..bbcfa2757c42945ed551646e75b75dfe576ec22f
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/goodness_chi_pvalue.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/goodness_summary.png b/radiant.basics/inst/app/tools/help/figures/goodness_summary.png
new file mode 100644
index 0000000000000000000000000000000000000000..056d580ddb7f0ca98225620aae6f5e47faa77e80
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/goodness_summary.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/prob_calc_batteries.png b/radiant.basics/inst/app/tools/help/figures/prob_calc_batteries.png
new file mode 100644
index 0000000000000000000000000000000000000000..638bb5a37e056c0b2592b0390656fc51816f84ab
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/prob_calc_batteries.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/prob_calc_headphones.png b/radiant.basics/inst/app/tools/help/figures/prob_calc_headphones.png
new file mode 100644
index 0000000000000000000000000000000000000000..9d7485c3bed477d4e11561dbe1280eb7644f0699
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/prob_calc_headphones.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/prob_calc_icecream.png b/radiant.basics/inst/app/tools/help/figures/prob_calc_icecream.png
new file mode 100644
index 0000000000000000000000000000000000000000..8ffaf3c8c0ca38d9720bdf17303b865872e8f6ad
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/prob_calc_icecream.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/single_mean_plot.png b/radiant.basics/inst/app/tools/help/figures/single_mean_plot.png
new file mode 100644
index 0000000000000000000000000000000000000000..5a9762da3892bb78d4f29b57030188f5abf48f68
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/single_mean_plot.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/single_mean_prob_calc.png b/radiant.basics/inst/app/tools/help/figures/single_mean_prob_calc.png
new file mode 100644
index 0000000000000000000000000000000000000000..0fb9912a2d24150730cb2ff7e7b2eacf9d205002
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/single_mean_prob_calc.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/single_mean_summary.png b/radiant.basics/inst/app/tools/help/figures/single_mean_summary.png
new file mode 100644
index 0000000000000000000000000000000000000000..0449b74dada49d3f2d104b5504c4aee1517498ee
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/single_mean_summary.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/single_prop_prob_calc_p.png b/radiant.basics/inst/app/tools/help/figures/single_prop_prob_calc_p.png
new file mode 100644
index 0000000000000000000000000000000000000000..a4c9fe3fbdeb7a4e29e8b2487bd8db99a05be032
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/single_prop_prob_calc_p.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/single_prop_prob_calc_v.png b/radiant.basics/inst/app/tools/help/figures/single_prop_prob_calc_v.png
new file mode 100644
index 0000000000000000000000000000000000000000..7fe9f957276fd5c93b34482eeee2041f44098363
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/single_prop_prob_calc_v.png differ
diff --git a/radiant.basics/inst/app/tools/help/figures/single_prop_summary.png b/radiant.basics/inst/app/tools/help/figures/single_prop_summary.png
new file mode 100644
index 0000000000000000000000000000000000000000..672f7375d929398a1e2f320e7c6f3fd60efeafcb
Binary files /dev/null and b/radiant.basics/inst/app/tools/help/figures/single_prop_summary.png differ
diff --git a/radiant.basics/inst/app/tools/help/goodness.md b/radiant.basics/inst/app/tools/help/goodness.md
new file mode 100644
index 0000000000000000000000000000000000000000..97bff7d8e6ef121b01d90fa6632a881a78f07936
--- /dev/null
+++ b/radiant.basics/inst/app/tools/help/goodness.md
@@ -0,0 +1,46 @@
+> 拟合优度检验用于确定样本数据是否与假设的分布一致
+
+### 示例
+
+数据来自 580 名报纸读者的样本,这些读者表明了(1)他们最常阅读的报纸(《今日美国》或《华尔街日报》)和(2)他们的收入水平(低收入 vs. 高收入)。数据包含三个变量:受访者标识符(id)、受访者收入(高或低)以及受访者主要阅读的报纸(《今日美国》或《华尔街日报》)。
+
+收集这些数据是为了研究收入水平与报纸选择之间是否存在关系。为确保结果具有可推广性,样本必须能代表目标总体。已知在该总体中,《今日美国》读者的相对比例高于《华尔街日报》读者,两者比例分别应为 55% 和 45%。我们可以使用拟合优度检验来检验以下原假设和备择假设:
+
+- H0:《今日美国》和《华尔街日报》的阅读份额分别为 55% 和 45%
+- Ha:《今日美国》和《华尔街日报》的阅读份额不等于上述设定值
+
+如果基于可用样本不能拒绝原假设,则观察数据与假设的总体份额或概率之间存在 “良好拟合”。在 Radiant(基础 > 表格 > 拟合优度检验)中,选择 “报纸(Newspaper)” 作为分类变量。如果我们将 “概率(Probabilities)” 输入框留空(或输入 1/2),则会检验份额是否相等。但为了检验 H0 和 Ha,我们需要输入`0.45 and 0.55`,然后按回车键。首先,比较观察频数和期望频数。期望频数基于原假设成立(即与设定份额无偏差)计算,公式为总样本量 ×p,其中 p 是某个单元格的假设份额(或概率)。
+
+





usethis::use_course("https://www.dropbox.com/sh/zw1yuiw8hvs47uc/AABPo1BncYv_i2eZfHQ7dgwCa?dl=1")
+
+Describing the Distribution of a Discrete Random
+ Variable (#1)
+
+* This video shows how to summarize information about a discrete random variable using the probability calculator in Radiant
+* Topics List:
+ - Calculate the mean and variance for a discrete random variable by hand
+ - Calculate the mean, variance, and select probabilities for a discrete random variable in Radiant
+
+Describing Normal and Binomial Distributions in Radiant(#2)
+
+* This video shows how to summarize information about Normal and Binomial distributions using the probability calculator in Radiant
+* Topics List:
+ - Calculate probabilities of a random variable following a Normal distribution in Radiant
+ - Calculate probabilities of a random variable following a Binomial distribution by hand
+ - Calculate probabilities of a random variable following a Binomial distribution in Radiant
+
+Describing Uniform and Binomial Distributions in Radiant(#3)
+
+* This video shows how to summarize information about Uniform and Binomial distributions using the probability calculator in Radiant
+* Topics List:
+ - Calculate probabilities of a random variable following a Uniform distribution in Radiant
+ - Calculate probabilities of a random variable following a Binomial distribution in Radiant
+
+Providing Probability Bounds(#4)
+
+* This video demonstrates how to provide probability bounds in Radiant
+* Topics List:
+ - Use probabilities as input type
+ - Round up the cutoff value
diff --git a/radiant.basics/inst/app/tools/help/prob_calc.md b/radiant.basics/inst/app/tools/help/prob_calc.md
new file mode 100644
index 0000000000000000000000000000000000000000..d98e2b9670b7c304f0c17b23ddc7181bf6b8f39b
--- /dev/null
+++ b/radiant.basics/inst/app/tools/help/prob_calc.md
@@ -0,0 +1,105 @@
+> 概率计算器
+
+基于二项分布(Binomial)、卡方分布(Chi-squared)、离散分布(Discrete)、F 分布(F)、指数分布(Exponential)、正态分布(Normal)、泊松分布(Poisson)、t 分布(t)或均匀分布(Uniform)计算概率或数值。
+
+## 电池测试
+
+假设《消费者报告》(CR)想要测试制造商关于电池寿命的声明。制造商声称,超过 90% 的电池可为笔记本电脑提供至少 12 小时的连续使用电力。CR 为 20 台相同的笔记本电脑配备了该制造商的电池。如果制造商的声明准确,那么 15 台或更多笔记本电脑在 12 小时后仍能运行的概率是多少?
+
+问题描述表明我们应从 “分布(Distribution)” 下拉菜单中选择 “二项分布(Binomial)”。要计算概率,选择 “数值(Values)” 作为 “输入类型(Input type)”,并输入`15`作为 “上限(Upper bound)”。在下方输出中,我们可以看到该概率为 0.989。恰好 15 台笔记本电脑在 12 小时后仍能运行的概率为 0.032。
+
+


usethis::use_course("https://www.dropbox.com/sh/zw1yuiw8hvs47uc/AABPo1BncYv_i2eZfHQ7dgwCa?dl=1")
+
+描述离散随机变量的分布(一)
+
+- 本视频展示如何使用 Radiant 中的概率计算器汇总离散随机变量的信息
+- 主题列表:
+ - 手动计算离散随机变量的均值和方差
+ - 在 Radiant 中计算离散随机变量的均值、方差和特定概率
+
+在 Radiant 中描述正态分布和二项分布(二)
+
+- 本视频展示如何使用 Radiant 中的概率计算器汇总正态分布和二项分布的信息
+- 主题列表:
+ - 在 Radiant 中计算服从正态分布的随机变量的概率
+ - 手动计算服从二项分布的随机变量的概率
+ - 在 Radiant 中计算服从二项分布的随机变量的概率
+
+在 Radiant 中描述均匀分布和二项分布(三)
+
+- 本视频展示如何使用 Radiant 中的概率计算器汇总均匀分布和二项分布的信息
+- 主题列表:
+ - 在 Radiant 中计算服从均匀分布的随机变量的概率
+ - 在 Radiant 中计算服从二项分布的随机变量的概率
+
+设置概率边界(四)
+
+- 本视频演示如何在 Radiant 中设置概率边界
+- 主题列表:
+ - 使用概率作为输入类型
+ - 对临界值向上取整
diff --git a/radiant.basics/inst/app/tools/help/single_mean.md b/radiant.basics/inst/app/tools/help/single_mean.md
new file mode 100644
index 0000000000000000000000000000000000000000..0aa97440684706a576fe4902bffc4de1821ea478
--- /dev/null
+++ b/radiant.basics/inst/app/tools/help/single_mean.md
@@ -0,0 +1,82 @@
+> 将单个均值与总体均值进行比较
+
+单样本均值 t 检验(或单样本 t 检验)用于将样本数据中某个变量的均值与我们样本数据所来自的总体中的(假设)均值进行比较。这很重要,因为我们很少能获取整个总体的数据。“比较值(Comparison value)” 框中指定了总体中的假设值。
+
+我们可以执行单侧检验(即`小于`或`大于`)或双侧检验(见 “备择假设(Alternative hypothesis)” 下拉菜单)。单侧检验用于评估现有数据是否提供证据表明样本均值大于(或小于)比较值(即原假设中的总体值)。
+
+## 示例
+
+我们获取了英国杂货店随机样本的数据。如果该产品类别的消费者需求超过 1 亿单位(即每家店约 1750 单位),管理层将考虑进入该市场。样本中每家店的平均需求为 1953 单位。虽然这个数字大于 1750,但我们需要确定这种差异是否可能由抽样误差导致。
+
+你可以在**demand_uk.rda**数据集中找到各样本店的单位销售量信息。该数据集包含两个变量:`store_id`(店铺 ID)和`demand_uk`(英国需求)。我们的原假设是英国每家店的平均需求等于 1750 单位,因此将该数值输入 “比较值(Comparison value)” 框。我们从 “备择假设(Alternative hypothesis)” 下拉菜单中选择 “大于(Greater than)” 选项,因为我们想确定现有数据是否提供足够证据拒绝原假设,支持英国每家店的平均需求**大于 1750 单位**的备择假设。
+
+


usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+单样本均值假设检验
+
+- 本视频展示如何检验关于单样本均值与总体均值的假设
+- 主题列表:
+ - 计算样本的汇总统计量
+ - 在 Radiant 中设置单样本均值的假设检验
+ - 使用 p 值、置信区间或临界值评估假设检验
diff --git a/radiant.basics/inst/app/tools/help/single_prop.md b/radiant.basics/inst/app/tools/help/single_prop.md
new file mode 100644
index 0000000000000000000000000000000000000000..79ce589c5b2f7a4f129735e810926f3d3435deec
--- /dev/null
+++ b/radiant.basics/inst/app/tools/help/single_prop.md
@@ -0,0 +1,87 @@
+> 将单个比例与总体比例进行比较
+
+单样本比例检验(或单样本二项检验)用于将样本数据中某类响应或取值的比例与我们样本数据所来自的总体中的(假设)比例进行比较。这很重要,因为我们很少能获取整个总体的数据。“比较值(Comparison value)” 框中指定了总体中的假设值。
+
+我们可以执行单侧检验(即`小于`或`大于`)或双侧检验(见 “备择假设(Alternative hypothesis)” 下拉菜单)。单侧检验用于评估现有数据是否提供证据表明样本比例大于(或小于)比较值(即原假设中的总体值)。
+
+## 示例
+
+一家汽车制造商通过在新目标市场随机抽样并采访 1000 名消费者开展了一项研究。研究目标是确定消费者是否会考虑购买该品牌汽车。
+
+管理层已决定公司将进入该细分市场。但如果品牌偏好率低于 10%,将投入额外资源用于广告和赞助,以提高目标消费者中的品牌知名度。在样本中,有 93 名消费者表现出公司所认为的强烈品牌喜爱度。
+
+你可以在**consider.rda**数据集中找到调查参与者的响应信息。该数据集包含两个变量:`id`和`consider`。
+
+我们的原假设是,会考虑在未来购买该汽车品牌的消费者比例等于 10%。从 “变量(Variable)” 下拉菜单中选择`consider`变量。要评估样本中`yes`响应的比例,从 “选择水平(Choose level)” 下拉菜单中选择`yes`。
+
+从 “备择假设(Alternative hypothesis)” 下拉菜单中选择 “小于(Less than)” 选项,以确定现有数据是否提供足够证据拒绝原假设,支持 “会考虑该品牌的消费者比例**小于 10%**” 的备择假设。
+
+


usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+单样本比例假设检验
+
+- 本视频展示如何检验关于单样本比例与总体比例的假设
+- 主题列表:
+ - 在 Radiant 中设置单样本比例的假设检验
+ - 使用 p 值、置信区间或临界值评估假设检验
diff --git a/radiant.basics/inst/app/ui.R b/radiant.basics/inst/app/ui.R
new file mode 100644
index 0000000000000000000000000000000000000000..3b532d0e954e66cbf682cc7859682a9db4f1e44e
--- /dev/null
+++ b/radiant.basics/inst/app/ui.R
@@ -0,0 +1,13 @@
+## ui for basics menu in radiant
+navbar_proj(
+ do.call(
+ navbarPage,
+ c(
+ "Radiant for R",
+ getOption("radiant.nav_ui"),
+ getOption("radiant.basics_ui"),
+ getOption("radiant.shared_ui"),
+ help_menu("help_basics_ui")
+ )
+ )
+)
diff --git a/radiant.basics/inst/app/www/js/run_return.js b/radiant.basics/inst/app/www/js/run_return.js
new file mode 100644
index 0000000000000000000000000000000000000000..8c79126445cd0cd7fa4e5c12da462b8eca9e9ea3
--- /dev/null
+++ b/radiant.basics/inst/app/www/js/run_return.js
@@ -0,0 +1,5 @@
+$(document).keydown(function(event) {
+ if ($("#cor_name").is(":focus") && event.keyCode == 13) {
+ $("#cor_store").click();
+ }
+});
diff --git a/radiant.basics/inst/translations/translation_zh.csv b/radiant.basics/inst/translations/translation_zh.csv
new file mode 100644
index 0000000000000000000000000000000000000000..a29bf6a2e8e532c5c23bde7c4b9e56da8e11dc6a
--- /dev/null
+++ b/radiant.basics/inst/translations/translation_zh.csv
@@ -0,0 +1,220 @@
+en,zh,source
+Help,帮助,"global.R, radiant.R"
+Keyboard shortcuts,键盘快捷键,global.R
+Normal,正态分布,clt_ui.R
+Binomial,二项分布,clt_ui.R
+Uniform,均匀分布,clt_ui.R
+Exponential,指数分布,clt_ui.R
+Sum,求和,clt_ui.R
+Mean,平均值,clt_ui.R
+Run simulation,运行模拟,clt_ui.R
+Re-run simulation,重新运行模拟,clt_ui.R
+Distribution:,分布:,clt_ui.R
+Min:,最小值:,clt_ui.R
+Max:,最大值:,clt_ui.R
+Mean:,均值:,clt_ui.R
+SD:,标准差:,clt_ui.R
+Rate:,速率:,clt_ui.R
+Size:,样本量:,clt_ui.R
+Prob:,概率:,clt_ui.R
+Sample size:,样本大小:,clt_ui.R
+# of samples:,样本数量:,clt_ui.R
+Number of bins:,分箱数量:,clt_ui.R
+Central Limit Theorem,中心极限定理,clt_ui.R
+Basics > Probability,基础 > 概率,clt_ui.R
+Please choose a sample size larger than 2,请选择一个大于 2 的样本大小,clt_ui.R
+Please choose 2 or more samples,请选择 2 个或更多样本,clt_ui.R
+Please choose a minimum value for the uniform distribution,请为均匀分布选择一个最小值,clt_ui.R
+Please choose a maximum value for the uniform distribution,请为均匀分布选择一个最大值,clt_ui.R
+The maximum value for the uniform distribution\nmust be larger than the minimum value,均匀分布的最大值必须大于最小值,clt_ui.R
+Please choose a mean value for the normal distribution,请为正态分布选择一个均值,clt_ui.R
+Please choose a non-zero standard deviation for the normal distribution,请为正态分布选择一个非零的标准差,clt_ui.R
+Please choose a rate larger than 1 for the exponential distribution,请为指数分布选择一个大于 1 的速率,clt_ui.R
+Please choose a size parameter larger than 1 for the binomial distribution,请为二项分布选择一个大于 1 的大小参数,clt_ui.R
+Please choose a probability between 0 and 1 for the binomial distribution,请为二项分布选择一个介于 0 到 1 之间的概率,clt_ui.R
+** Press the Run simulation button to simulate data **,** 点击运行模拟按钮以生成数据 **,clt_ui.R
+Generating plots,正在生成图形,"clt_ui.R, compare_means_ui.R"
+Save central limit theorem plot,保存中心极限定理图,clt_ui.R
+Two sided,双侧,compare_means_ui.R
+Less than,小于,compare_means_ui.R
+Greater than,大于,compare_means_ui.R
+independent,独立样本,compare_means_ui.R
+paired,配对样本,compare_means_ui.R
+None,无,compare_means_ui.R
+Bonferroni,Bonferroni 校正,compare_means_ui.R
+Scatter,散点图,compare_means_ui.R
+Box,箱线图,compare_means_ui.R
+Density,密度图,compare_means_ui.R
+Bar,条形图,compare_means_ui.R
+Select a factor or numeric variable:,选择一个因子或数值变量:,compare_means_ui.R
+Numeric variable(s):,数值变量(可多选):,compare_means_ui.R
+Numeric variable:,数值变量:,compare_means_ui.R
+Choose combinations:,选择组合:,compare_means_ui.R
+Evaluate all combinations,评估所有组合,compare_means_ui.R
+Alternative hypothesis:,备择假设:,compare_means_ui.R
+Confidence level:,置信水平:,compare_means_ui.R
+Show additional statistics,显示额外统计量,compare_means_ui.R
+Sample type:,样本类型:,compare_means_ui.R
+Multiple comp. adjustment:,多重比较校正:,compare_means_ui.R
+Test type:,检验类型:,compare_means_ui.R
+t-test,t 检验,compare_means_ui.R
+Wilcox,Wilcoxon 检验,compare_means_ui.R
+Select plots:,选择绘图类型:,compare_means_ui.R
+Select plots,选择绘图,compare_means_ui.R
+Compare means,均值比较,compare_means_ui.R
+Summary,摘要,compare_means_ui.R
+Plot,图形,compare_means_ui.R
+Basics > Means,基础 > 均值,compare_means_ui.R
+"This analysis requires at least two variables. The first can be of type
+factor, numeric, or interval. The second must be of type numeric or interval.
+If these variable types are not available please select another dataset.
+
+","该分析至少需要两个变量。
+第一个变量可以是因子、数值或区间类型,第二个变量必须是数值或区间类型。
+如果这些类型的变量不可用,请选择其他数据集。
+
+",compare_means_ui.R
+Nothing to plot. Please select a plot type,没有可绘制的内容,请选择绘图类型,compare_means_ui.R
+Save compare means plot,保存均值比较图,compare_means_ui.R
+Basics > Proportions,基础 > 比例,compare_props_ui.R
+Compare proportions,比较比例,compare_props_ui.R
+"This analysis requires two categorical variables. The first must have
+two or more levels. The second can have only two levels. If these
+variable types are not available please select another dataset.
+
+","该分析需要两个分类变量。
+第一个变量必须具有两个或更多水平,第二个变量只能有两个水平。
+如果这些变量类型不可用,请选择其他数据集。
+
+",compare_props_ui.R
+Select a grouping variable:,选择分组变量:,compare_props_ui.R
+Save compare proportions plot,保存比较比例图,compare_props_ui.R
+Dodge,并列柱状图,compare_props_ui.R
+Variable (select one):,变量(选择一个):,compare_props_ui.R
+Pearson,皮尔逊积矩相关,correlation_ui.R
+Spearman,斯皮尔曼秩相关,correlation_ui.R
+Kendall,肯德尔秩相关,correlation_ui.R
+Calculate correlation,计算相关性,correlation_ui.R
+Basics > Tables,基础 > 表格,correlation_ui.R
+Correlation,相关性,correlation_ui.R
+Adjust for {factor} variables,针对 {factor} 变量进行调整,correlation_ui.R
+Calculate adjusted p.values,计算调整后的 p 值,correlation_ui.R
+Correlation cutoff:,相关性阈值:,correlation_ui.R
+Show covariance matrix,显示协方差矩阵,correlation_ui.R
+Store,存储,correlation_ui.R
+"This analysis requires two or more variables or type numeric, integer,or date. If these variable types are not available please select another dataset.",该分析需要两个或以上的数值型、整数型或日期型变量。如果这些变量类型不可用,请选择其他数据集。,correlation_ui.R
+Method:,方法:,correlation_ui.R
+Acquiring variable information,正在获取变量信息,correlation_ui.R
+Select variables:,选择变量:,correlation_ui.R
+Store as data.frame:,存储为数据框:,correlation_ui.R
+"This analysis requires two or more variables or type numeric,\ninteger,or date. If these variable types are not available\nplease select another dataset.\n\n",本分析需要两个或以上的变量,并且类型必须是数值型、整数型或日期型。如果这些变量类型不可用,请选择其他数据集。,correlation_ui.R
+Save correlation plot,保存相关性图表,correlation_ui.R
+Number of data points plotted:,绘制的数据点数量:,correlation_ui.R
+"This analysis requires two or more variables or type numeric,
+integer,or date. If these variable types are not available
+please select another dataset.
+
+","本分析需要两个或以上的变量,并且类型必须是数值型、整数型或日期型。
+如果这些变量类型不可用,请选择其他数据集。
+
+",correlation_ui.R
+Re-calculate correlations,重新计算相关系数,correlation_ui.R
+Observed,观察值,cross_tabs_ui.R
+Expected,期望值,cross_tabs_ui.R
+Chi-squared,卡方值,cross_tabs_ui.R
+Deviation std.,标准差偏差,cross_tabs_ui.R
+Row percentages,行百分比,cross_tabs_ui.R
+Column percentages,列百分比,cross_tabs_ui.R
+Table percentages,表格百分比,cross_tabs_ui.R
+Cross-tabs,交叉表,cross_tabs_ui.R
+"This analysis requires two categorical variables. Both must have two or more levels.
+If these variable types are not available please select another dataset.
+
+","此分析需要两个分类变量,且每个变量必须至少有两个水平。
+如果这些类型的变量不可用,请选择其他数据集。
+
+",cross_tabs_ui.R
+Select a categorical variable:,请选择一个分类变量:,cross_tabs_ui.R
+Save cross-tabs plot,保存交叉表图形,cross_tabs_ui.R
+Goodness of fit,拟合优度检验,goodness_ui.R
+"This analysis requires a categorical variables with two or more levels.
+If such a variable type is not available please select another dataset.
+
+","此分析需要一个具有两个或以上水平的分类变量。
+如果没有这种类型的变量,请选择其他数据集。
+
+",goodness_ui.R
+Save goodness of fit plot,保存拟合优度检验图形,goodness_ui.R
+Probabilities:,概率:,goodness_ui.R
+"Enter probabilities (e.g., 1/2 1/2)",输入概率(例如:1/2 1/2),goodness_ui.R
+Discrete,离散分布,prob_calc_ui.R
+F,F 分布,prob_calc_ui.R
+Log normal,对数正态,prob_calc_ui.R
+Poisson,泊松分布,prob_calc_ui.R
+Values,数值,prob_calc_ui.R
+Probability calculator,概率计算器,prob_calc_ui.R
+Input type:,输入类型:,prob_calc_ui.R
+Decimals:,小数位数:,prob_calc_ui.R
+Save probability calculator plot,保存概率计算器图形,prob_calc_ui.R
+Please provide a mean and standard deviation (> 0),请提供平均值和标准差(标准差需大于 0),prob_calc_ui.R
+St. dev:,标准差:,prob_calc_ui.R
+Lower bound:,下限:,prob_calc_ui.R
+Upper bound:,上限:,prob_calc_ui.R
+Provide an integer value for the number of decimal places,请输入整数,表示保留的小数位数,prob_calc_ui.R
+"Please provide a set of values and probabilities.
+Separate numbers using spaces (e.g., 1/2 1/2)","请提供一组数值和对应的概率。
+请用空格分隔(例如:1/2 1/2)",prob_calc_ui.R
+Values:,数值:,prob_calc_ui.R
+Please provide a value for n (number of trials) and p (probability of success),请提供试验次数 (n) 和成功概率 (p) 的值,prob_calc_ui.R
+Please provide a minimum and a maximum value,请提供最小值和最大值,prob_calc_ui.R
+Please provide a value for the degrees of freedom (> 0),请提供大于 0 的自由度值,prob_calc_ui.R
+"Please provide a value for Degrees of freedom 1 (> 0)
+and for Degrees of freedom 2 (> 4)",请提供自由度 1(大于 0)和自由度 2(大于 4)的值,prob_calc_ui.R
+Please provide a value for the rate (> 0),请提供大于 0 的速率值,prob_calc_ui.R
+Please provide a value for lambda (> 0),请提供大于 0 的 λ(Lambda)值,prob_calc_ui.R
+n:,试验次数:,prob_calc_ui.R
+p:,成功概率:,prob_calc_ui.R
+Degrees of freedom:,自由度:,prob_calc_ui.R
+Degrees of freedom 1:,自由度 1:,prob_calc_ui.R
+Degrees of freedom 2:,自由度 2:,prob_calc_ui.R
+Mean log:,对数均值:,prob_calc_ui.R
+St. dev log:,对数标准差:,prob_calc_ui.R
+Lambda:,λ:,prob_calc_ui.R
+Histogram,直方图,single_mean_ui.R
+Simulate,模拟,single_mean_ui.R
+Single mean,单样本均值,single_mean_ui.R
+Comparison value:,比较值:,single_mean_ui.R
+"This analysis requires a variable of type numeric or interval. If none are
+available please select another dataset.
+
+","此分析需要一个数值型或区间型变量。
+如果当前数据集中没有此类变量,请选择其他数据集。
+
+",single_mean_ui.R
+Save single mean plot,保存单样本均值图表,single_mean_ui.R
+Single proportion,单样本比例,single_prop_ui.R
+Binomial exact,精确二项检验,single_prop_ui.R
+Z-test,Z 检验,single_prop_ui.R
+Choose level:,选择水平:,single_prop_ui.R
+"This analysis requires a categorical variable. In none are available
+please select another dataset.
+
+","本分析需要一个分类变量。
+如果没有可用的分类变量,请选择其他数据集。
+
+",single_prop_ui.R
+Save single proportion plot,保存单样本比例图,single_prop_ui.R
+Basics,基础,init.R
+Probability,概率,init.R
+Probability calculator,概率计算器,init.R
+Central Limit Theorem,中心极限定理,init.R
+Means,均值,init.R
+Single mean,单样本均值,init.R
+Compare means,均值比较,init.R
+Proportions,比例,init.R
+Single proportion,单样本比例,init.R
+Compare proportions,比例比较,init.R
+Tables,表格,init.R
+Goodness of fit,拟合优度,init.R
+Cross-tabs,交叉表,init.R
+Correlation,相关性,init.R
diff --git a/radiant.basics/man/clt.Rd b/radiant.basics/man/clt.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..818c961a7b7bff10163b58af0c90df128b03f496
--- /dev/null
+++ b/radiant.basics/man/clt.Rd
@@ -0,0 +1,53 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/clt.R
+\name{clt}
+\alias{clt}
+\title{Central Limit Theorem simulation}
+\usage{
+clt(
+ dist,
+ n = 100,
+ m = 100,
+ norm_mean = 0,
+ norm_sd = 1,
+ binom_size = 10,
+ binom_prob = 0.2,
+ unif_min = 0,
+ unif_max = 1,
+ expo_rate = 1
+)
+}
+\arguments{
+\item{dist}{Distribution to simulate}
+
+\item{n}{Sample size}
+
+\item{m}{Number of samples}
+
+\item{norm_mean}{Mean for the normal distribution}
+
+\item{norm_sd}{Standard deviation for the normal distribution}
+
+\item{binom_size}{Size for the binomial distribution}
+
+\item{binom_prob}{Probability for the binomial distribution}
+
+\item{unif_min}{Minimum for the uniform distribution}
+
+\item{unif_max}{Maximum for the uniform distribution}
+
+\item{expo_rate}{Rate for the exponential distribution}
+}
+\value{
+A list with the name of the Distribution and a matrix of simulated data
+}
+\description{
+Central Limit Theorem simulation
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/clt.html} for an example in Radiant
+}
+\examples{
+clt("Uniform", 10, 10, unif_min = 10, unif_max = 20)
+
+}
diff --git a/radiant.basics/man/compare_means.Rd b/radiant.basics/man/compare_means.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..399cf1610384e0b3c4c7bd397175aa5ef6a29118
--- /dev/null
+++ b/radiant.basics/man/compare_means.Rd
@@ -0,0 +1,61 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/compare_means.R
+\name{compare_means}
+\alias{compare_means}
+\title{Compare sample means}
+\usage{
+compare_means(
+ dataset,
+ var1,
+ var2,
+ samples = "independent",
+ alternative = "two.sided",
+ conf_lev = 0.95,
+ comb = "",
+ adjust = "none",
+ test = "t",
+ data_filter = "",
+ envir = parent.frame()
+)
+}
+\arguments{
+\item{dataset}{Dataset}
+
+\item{var1}{A numeric variable or factor selected for comparison}
+
+\item{var2}{One or more numeric variables for comparison. If var1 is a factor only one variable can be selected and the mean of this variable is compared across (factor) levels of var1}
+
+\item{samples}{Are samples independent ("independent") or not ("paired")}
+
+\item{alternative}{The alternative hypothesis ("two.sided", "greater" or "less")}
+
+\item{conf_lev}{Span of the confidence interval}
+
+\item{comb}{Combinations to evaluate}
+
+\item{adjust}{Adjustment for multiple comparisons ("none" or "bonf" for Bonferroni)}
+
+\item{test}{t-test ("t") or Wilcox ("wilcox")}
+
+\item{data_filter}{Expression entered in, e.g., Data > View to filter the dataset in Radiant. The expression should be a string (e.g., "price > 10000")}
+
+\item{envir}{Environment to extract data from}
+}
+\value{
+A list of all variables defined in the function as an object of class compare_means
+}
+\description{
+Compare sample means
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/compare_means.html} for an example in Radiant
+}
+\examples{
+compare_means(diamonds, "cut", "price") \%>\% str()
+
+}
+\seealso{
+\code{\link{summary.compare_means}} to summarize results
+
+\code{\link{plot.compare_means}} to plot results
+}
diff --git a/radiant.basics/man/compare_props.Rd b/radiant.basics/man/compare_props.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..d4e1cdf2eec88fdbe9f02f2e725ca3780cba2adc
--- /dev/null
+++ b/radiant.basics/man/compare_props.Rd
@@ -0,0 +1,58 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/compare_props.R
+\name{compare_props}
+\alias{compare_props}
+\title{Compare sample proportions across groups}
+\usage{
+compare_props(
+ dataset,
+ var1,
+ var2,
+ levs = "",
+ alternative = "two.sided",
+ conf_lev = 0.95,
+ comb = "",
+ adjust = "none",
+ data_filter = "",
+ envir = parent.frame()
+)
+}
+\arguments{
+\item{dataset}{Dataset}
+
+\item{var1}{A grouping variable to split the data for comparisons}
+
+\item{var2}{The variable to calculate proportions for}
+
+\item{levs}{The factor level selected for the proportion comparison}
+
+\item{alternative}{The alternative hypothesis ("two.sided", "greater" or "less")}
+
+\item{conf_lev}{Span of the confidence interval}
+
+\item{comb}{Combinations to evaluate}
+
+\item{adjust}{Adjustment for multiple comparisons ("none" or "bonf" for Bonferroni)}
+
+\item{data_filter}{Expression entered in, e.g., Data > View to filter the dataset in Radiant. The expression should be a string (e.g., "price > 10000")}
+
+\item{envir}{Environment to extract data from}
+}
+\value{
+A list of all variables defined in the function as an object of class compare_props
+}
+\description{
+Compare sample proportions across groups
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/compare_props.html} for an example in Radiant
+}
+\examples{
+compare_props(titanic, "pclass", "survived") \%>\% str()
+
+}
+\seealso{
+\code{\link{summary.compare_props}} to summarize results
+
+\code{\link{plot.compare_props}} to plot results
+}
diff --git a/radiant.basics/man/consider.Rd b/radiant.basics/man/consider.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..b340dfbef754283e2baf633a77414977536813c7
--- /dev/null
+++ b/radiant.basics/man/consider.Rd
@@ -0,0 +1,19 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/aaa.R
+\docType{data}
+\name{consider}
+\alias{consider}
+\title{Car brand consideration}
+\format{
+A data frame with 1000 rows and 2 variables
+}
+\usage{
+data(consider)
+}
+\description{
+Car brand consideration
+}
+\details{
+Survey data of consumer purchase intentions. Description provided in attr(consider,"description")
+}
+\keyword{datasets}
diff --git a/radiant.basics/man/cor2df.Rd b/radiant.basics/man/cor2df.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..f0e54e32d823dcb0920e9d709df2766d7a1b748b
--- /dev/null
+++ b/radiant.basics/man/cor2df.Rd
@@ -0,0 +1,21 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/correlation.R
+\name{cor2df}
+\alias{cor2df}
+\title{Store a correlation matrix as a (long) data.frame}
+\usage{
+cor2df(object, labels = c("label1", "label2"), ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{correlation}}}
+
+\item{labels}{Column names for the correlation pairs}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Store a correlation matrix as a (long) data.frame
+}
+\details{
+Return the correlation matrix as a (long) data.frame. See \url{https://radiant-rstats.github.io/docs/basics/correlation.html} for an example in Radiant
+}
diff --git a/radiant.basics/man/correlation.Rd b/radiant.basics/man/correlation.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..f3a4049cb3c6a8de27a9919c16fe044008a5690c
--- /dev/null
+++ b/radiant.basics/man/correlation.Rd
@@ -0,0 +1,50 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/correlation.R
+\name{correlation}
+\alias{correlation}
+\title{Calculate correlations for two or more variables}
+\usage{
+correlation(
+ dataset,
+ vars = "",
+ method = "pearson",
+ hcor = FALSE,
+ hcor_se = FALSE,
+ data_filter = "",
+ envir = parent.frame()
+)
+}
+\arguments{
+\item{dataset}{Dataset}
+
+\item{vars}{Variables to include in the analysis. Default is all but character and factor variables with more than two unique values are removed}
+
+\item{method}{Type of correlations to calculate. Options are "pearson", "spearman", and "kendall". "pearson" is the default}
+
+\item{hcor}{Use polycor::hetcor to calculate the correlation matrix}
+
+\item{hcor_se}{Calculate standard errors when using polycor::hetcor}
+
+\item{data_filter}{Expression entered in, e.g., Data > View to filter the dataset in Radiant. The expression should be a string (e.g., "price > 10000")}
+
+\item{envir}{Environment to extract data from}
+}
+\value{
+A list with all variables defined in the function as an object of class compare_means
+}
+\description{
+Calculate correlations for two or more variables
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/correlation.html} for an example in Radiant
+}
+\examples{
+correlation(diamonds, c("price", "carat")) \%>\% str()
+correlation(diamonds, "x:z") \%>\% str()
+
+}
+\seealso{
+\code{\link{summary.correlation}} to summarize results
+
+\code{\link{plot.correlation}} to plot results
+}
diff --git a/radiant.basics/man/cross_tabs.Rd b/radiant.basics/man/cross_tabs.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..1456c010c70770630d8b5f69757ed291dfb3b98d
--- /dev/null
+++ b/radiant.basics/man/cross_tabs.Rd
@@ -0,0 +1,47 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/cross_tabs.R
+\name{cross_tabs}
+\alias{cross_tabs}
+\title{Evaluate associations between categorical variables}
+\usage{
+cross_tabs(
+ dataset,
+ var1,
+ var2,
+ tab = NULL,
+ data_filter = "",
+ envir = parent.frame()
+)
+}
+\arguments{
+\item{dataset}{Dataset (i.e., a data.frame or table)}
+
+\item{var1}{A categorical variable}
+
+\item{var2}{A categorical variable}
+
+\item{tab}{Table with frequencies as alternative to dataset}
+
+\item{data_filter}{Expression entered in, e.g., Data > View to filter the dataset in Radiant. The expression should be a string (e.g., "price > 10000")}
+
+\item{envir}{Environment to extract data from}
+}
+\value{
+A list of all variables used in cross_tabs as an object of class cross_tabs
+}
+\description{
+Evaluate associations between categorical variables
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/cross_tabs.html} for an example in Radiant
+}
+\examples{
+cross_tabs(newspaper, "Income", "Newspaper") \%>\% str()
+table(select(newspaper, Income, Newspaper)) \%>\% cross_tabs(tab = .)
+
+}
+\seealso{
+\code{\link{summary.cross_tabs}} to summarize results
+
+\code{\link{plot.cross_tabs}} to plot results
+}
diff --git a/radiant.basics/man/demand_uk.Rd b/radiant.basics/man/demand_uk.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..ec4f9ea65ecdd9842cf2d837a5fc2386cdc06f65
--- /dev/null
+++ b/radiant.basics/man/demand_uk.Rd
@@ -0,0 +1,19 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/aaa.R
+\docType{data}
+\name{demand_uk}
+\alias{demand_uk}
+\title{Demand in the UK}
+\format{
+A data frame with 1000 rows and 2 variables
+}
+\usage{
+data(demand_uk)
+}
+\description{
+Demand in the UK
+}
+\details{
+Survey data of consumer purchase intentions. Description provided in attr(demand_uk,"description")
+}
+\keyword{datasets}
diff --git a/radiant.basics/man/goodness.Rd b/radiant.basics/man/goodness.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..d9599a766bc8c985d7f57b67fbc8f228fb68f227
--- /dev/null
+++ b/radiant.basics/man/goodness.Rd
@@ -0,0 +1,48 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/goodness.R
+\name{goodness}
+\alias{goodness}
+\title{Evaluate if sample data for a categorical variable is consistent with a hypothesized distribution}
+\usage{
+goodness(
+ dataset,
+ var,
+ p = NULL,
+ tab = NULL,
+ data_filter = "",
+ envir = parent.frame()
+)
+}
+\arguments{
+\item{dataset}{Dataset}
+
+\item{var}{A categorical variable}
+
+\item{p}{Hypothesized distribution as a number, fraction, or numeric vector. If unspecified, defaults to an even distribution}
+
+\item{tab}{Table with frequencies as alternative to dataset}
+
+\item{data_filter}{Expression entered in, e.g., Data > View to filter the dataset in Radiant. The expression should be a string (e.g., "price > 10000")}
+
+\item{envir}{Environment to extract data from}
+}
+\value{
+A list of all variables used in goodness as an object of class goodness
+}
+\description{
+Evaluate if sample data for a categorical variable is consistent with a hypothesized distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/goodness.html} for an example in Radiant
+}
+\examples{
+goodness(newspaper, "Income") \%>\% str()
+goodness(newspaper, "Income", p = c(3 / 4, 1 / 4)) \%>\% str()
+table(select(newspaper, Income)) \%>\% goodness(tab = .)
+
+}
+\seealso{
+\code{\link{summary.goodness}} to summarize results
+
+\code{\link{plot.goodness}} to plot results
+}
diff --git a/radiant.basics/man/newspaper.Rd b/radiant.basics/man/newspaper.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..755b4b98c00a44da355b682133536ae4a02616d8
--- /dev/null
+++ b/radiant.basics/man/newspaper.Rd
@@ -0,0 +1,19 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/aaa.R
+\docType{data}
+\name{newspaper}
+\alias{newspaper}
+\title{Newspaper readership}
+\format{
+A data frame with 580 rows and 2 variables
+}
+\usage{
+data(newspaper)
+}
+\description{
+Newspaper readership
+}
+\details{
+Newspaper readership data for 580 consumers. Description provided in attr(newspaper,"description")
+}
+\keyword{datasets}
diff --git a/radiant.basics/man/plot.clt.Rd b/radiant.basics/man/plot.clt.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..1bade81d1765d5f71062cacdcb92773137880d81
--- /dev/null
+++ b/radiant.basics/man/plot.clt.Rd
@@ -0,0 +1,27 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/clt.R
+\name{plot.clt}
+\alias{plot.clt}
+\title{Plot method for the Central Limit Theorem simulation}
+\usage{
+\method{plot}{clt}(x, stat = "sum", bins = 15, ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{clt}}}
+
+\item{stat}{Statistic to use (sum or mean)}
+
+\item{bins}{Number of bins to use}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the Central Limit Theorem simulation
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/clt.html} for an example in Radiant
+}
+\examples{
+clt("Uniform", 100, 100, unif_min = 10, unif_max = 20) \%>\% plot()
+
+}
diff --git a/radiant.basics/man/plot.compare_means.Rd b/radiant.basics/man/plot.compare_means.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..91d95b2e4245677998f0f742eb5289367c2d7262
--- /dev/null
+++ b/radiant.basics/man/plot.compare_means.Rd
@@ -0,0 +1,35 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/compare_means.R
+\name{plot.compare_means}
+\alias{plot.compare_means}
+\title{Plot method for the compare_means function}
+\usage{
+\method{plot}{compare_means}(x, plots = "scatter", shiny = FALSE, custom = FALSE, ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{compare_means}}}
+
+\item{plots}{One or more plots ("bar", "density", "box", or "scatter")}
+
+\item{shiny}{Did the function call originate inside a shiny app}
+
+\item{custom}{Logical (TRUE, FALSE) to indicate if ggplot object (or list of ggplot objects) should be returned. This option can be used to customize plots (e.g., add a title, change x and y labels, etc.). See examples and \url{https://ggplot2.tidyverse.org/} for options.}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the compare_means function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/compare_means.html} for an example in Radiant
+}
+\examples{
+result <- compare_means(diamonds, "cut", "price")
+plot(result, plots = c("bar", "density"))
+
+}
+\seealso{
+\code{\link{compare_means}} to calculate results
+
+\code{\link{summary.compare_means}} to summarize results
+}
diff --git a/radiant.basics/man/plot.compare_props.Rd b/radiant.basics/man/plot.compare_props.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..3ca4263d2da1c387c40f9ce1a71f0ad57d167cd4
--- /dev/null
+++ b/radiant.basics/man/plot.compare_props.Rd
@@ -0,0 +1,35 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/compare_props.R
+\name{plot.compare_props}
+\alias{plot.compare_props}
+\title{Plot method for the compare_props function}
+\usage{
+\method{plot}{compare_props}(x, plots = "bar", shiny = FALSE, custom = FALSE, ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{compare_props}}}
+
+\item{plots}{One or more plots of proportions ("bar" or "dodge")}
+
+\item{shiny}{Did the function call originate inside a shiny app}
+
+\item{custom}{Logical (TRUE, FALSE) to indicate if ggplot object (or list of ggplot objects) should be returned. This option can be used to customize plots (e.g., add a title, change x and y labels, etc.). See examples and \url{https://ggplot2.tidyverse.org/} for options.}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the compare_props function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/compare_props.html} for an example in Radiant
+}
+\examples{
+result <- compare_props(titanic, "pclass", "survived")
+plot(result, plots = c("bar", "dodge"))
+
+}
+\seealso{
+\code{\link{compare_props}} to calculate results
+
+\code{\link{summary.compare_props}} to summarize results
+}
diff --git a/radiant.basics/man/plot.correlation.Rd b/radiant.basics/man/plot.correlation.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..0a1178ef38e88922d9d2d4ea20b93e464ead3d97
--- /dev/null
+++ b/radiant.basics/man/plot.correlation.Rd
@@ -0,0 +1,35 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/correlation.R
+\name{plot.correlation}
+\alias{plot.correlation}
+\title{Plot method for the correlation function}
+\usage{
+\method{plot}{correlation}(x, nrobs = -1, jit = c(0, 0), dec = 2, ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{correlation}}}
+
+\item{nrobs}{Number of data points to show in scatter plots (-1 for all)}
+
+\item{jit}{A numeric vector that determines the amount of jittering to apply to the x and y variables in a scatter plot. Default is 0. Use, e.g., 0.3 to add some jittering}
+
+\item{dec}{Number of decimals to show}
+
+\item{...}{further arguments passed to or from other methods.}
+}
+\description{
+Plot method for the correlation function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/correlation.html} for an example in Radiant
+}
+\examples{
+result <- correlation(diamonds, c("price", "carat", "table"))
+plot(result)
+
+}
+\seealso{
+\code{\link{correlation}} to calculate results
+
+\code{\link{summary.correlation}} to summarize results
+}
diff --git a/radiant.basics/man/plot.cross_tabs.Rd b/radiant.basics/man/plot.cross_tabs.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..f473abd01fc2ac5489f8a80a7b6cb1ab1b4d3b3b
--- /dev/null
+++ b/radiant.basics/man/plot.cross_tabs.Rd
@@ -0,0 +1,35 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/cross_tabs.R
+\name{plot.cross_tabs}
+\alias{plot.cross_tabs}
+\title{Plot method for the cross_tabs function}
+\usage{
+\method{plot}{cross_tabs}(x, check = "", shiny = FALSE, custom = FALSE, ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{cross_tabs}}}
+
+\item{check}{Show plots for variables var1 and var2. "observed" for the observed frequencies table, "expected" for the expected frequencies table (i.e., frequencies that would be expected if the null hypothesis holds), "chi_sq" for the contribution to the overall chi-squared statistic for each cell (i.e., (o - e)^2 / e), "dev_std" for the standardized differences between the observed and expected frequencies (i.e., (o - e) / sqrt(e)), and "row_perc", "col_perc", and "perc" for row, column, and table percentages respectively}
+
+\item{shiny}{Did the function call originate inside a shiny app}
+
+\item{custom}{Logical (TRUE, FALSE) to indicate if ggplot object (or list of ggplot objects) should be returned. This option can be used to customize plots (e.g., add a title, change x and y labels, etc.). See examples and \url{https://ggplot2.tidyverse.org/} for options.}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the cross_tabs function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/cross_tabs.html} for an example in Radiant
+}
+\examples{
+result <- cross_tabs(newspaper, "Income", "Newspaper")
+plot(result, check = c("observed", "expected", "chi_sq"))
+
+}
+\seealso{
+\code{\link{cross_tabs}} to calculate results
+
+\code{\link{summary.cross_tabs}} to summarize results
+}
diff --git a/radiant.basics/man/plot.goodness.Rd b/radiant.basics/man/plot.goodness.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..cf51cf4490117e9116ac657147cc8085efa5308a
--- /dev/null
+++ b/radiant.basics/man/plot.goodness.Rd
@@ -0,0 +1,38 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/goodness.R
+\name{plot.goodness}
+\alias{plot.goodness}
+\title{Plot method for the goodness function}
+\usage{
+\method{plot}{goodness}(x, check = "", fillcol = "blue", shiny = FALSE, custom = FALSE, ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{goodness}}}
+
+\item{check}{Show plots for variable var. "observed" for the observed frequencies table, "expected" for the expected frequencies table (i.e., frequencies that would be expected if the null hypothesis holds), "chi_sq" for the contribution to the overall chi-squared statistic for each cell (i.e., (o - e)^2 / e), and "dev_std" for the standardized differences between the observed and expected frequencies (i.e., (o - e) / sqrt(e))}
+
+\item{fillcol}{Color used for bar plots}
+
+\item{shiny}{Did the function call originate inside a shiny app}
+
+\item{custom}{Logical (TRUE, FALSE) to indicate if ggplot object (or list of ggplot objects) should be returned. This option can be used to customize plots (e.g., add a title, change x and y labels, etc.). See examples and \url{https://ggplot2.tidyverse.org/} for options.}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the goodness function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/goodness} for an example in Radiant
+}
+\examples{
+result <- goodness(newspaper, "Income")
+plot(result, check = c("observed", "expected", "chi_sq"))
+goodness(newspaper, "Income") \%>\% plot(c("observed", "expected"))
+
+}
+\seealso{
+\code{\link{goodness}} to calculate results
+
+\code{\link{summary.goodness}} to summarize results
+}
diff --git a/radiant.basics/man/plot.prob_binom.Rd b/radiant.basics/man/plot.prob_binom.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..a68c582640bc4d69b0a735a300f60f922bba8969
--- /dev/null
+++ b/radiant.basics/man/plot.prob_binom.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_binom}
+\alias{plot.prob_binom}
+\title{Plot method for the probability calculator (binomial)}
+\usage{
+\method{plot}{prob_binom}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_binom}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (binomial)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_binom(n = 10, p = 0.3, ub = 3)
+plot(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_binom}} to calculate results
+
+\code{\link{summary.prob_binom}} to summarize results
+}
diff --git a/radiant.basics/man/plot.prob_chisq.Rd b/radiant.basics/man/plot.prob_chisq.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..ba4e037657b1fb4d18d8f1470218d8babbac9842
--- /dev/null
+++ b/radiant.basics/man/plot.prob_chisq.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_chisq}
+\alias{plot.prob_chisq}
+\title{Plot method for the probability calculator (Chi-squared distribution)}
+\usage{
+\method{plot}{prob_chisq}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_chisq}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (Chi-squared distribution)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_chisq(df = 1, ub = 3.841)
+plot(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_chisq}} to calculate results
+
+\code{\link{summary.prob_chisq}} to summarize results
+}
diff --git a/radiant.basics/man/plot.prob_disc.Rd b/radiant.basics/man/plot.prob_disc.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..10f3ff4d79c4ec404235211bc2edc427c08fdd15
--- /dev/null
+++ b/radiant.basics/man/plot.prob_disc.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_disc}
+\alias{plot.prob_disc}
+\title{Plot method for the probability calculator (discrete)}
+\usage{
+\method{plot}{prob_disc}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_disc}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (discrete)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_disc(v = 1:6, p = c(2 / 6, 2 / 6, 1 / 12, 1 / 12, 1 / 12, 1 / 12), pub = 0.95)
+plot(result, type = "probs")
+
+}
+\seealso{
+\code{\link{prob_disc}} to calculate results
+
+\code{\link{summary.prob_disc}} to summarize results
+}
diff --git a/radiant.basics/man/plot.prob_expo.Rd b/radiant.basics/man/plot.prob_expo.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..f6453aaabb83a484e5280c87280a4ff1fa130250
--- /dev/null
+++ b/radiant.basics/man/plot.prob_expo.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_expo}
+\alias{plot.prob_expo}
+\title{Plot method for the probability calculator (Exponential distribution)}
+\usage{
+\method{plot}{prob_expo}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_expo}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (Exponential distribution)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_expo(rate = 1, ub = 2.996)
+plot(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_expo}} to calculate results
+
+\code{\link{summary.prob_expo}} to summarize results
+}
diff --git a/radiant.basics/man/plot.prob_fdist.Rd b/radiant.basics/man/plot.prob_fdist.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..3fbbfaa998afdd13a7db9013ece65582f85a49a8
--- /dev/null
+++ b/radiant.basics/man/plot.prob_fdist.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_fdist}
+\alias{plot.prob_fdist}
+\title{Plot method for the probability calculator (F-distribution)}
+\usage{
+\method{plot}{prob_fdist}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_fdist}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (F-distribution)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_fdist(df1 = 10, df2 = 10, ub = 2.978)
+plot(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_fdist}} to calculate results
+
+\code{\link{summary.prob_fdist}} to summarize results
+}
diff --git a/radiant.basics/man/plot.prob_lnorm.Rd b/radiant.basics/man/plot.prob_lnorm.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..e5335aeb6f9ad26ced2832d3840c18d3ccad46d2
--- /dev/null
+++ b/radiant.basics/man/plot.prob_lnorm.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_lnorm}
+\alias{plot.prob_lnorm}
+\title{Plot method for the probability calculator (log normal)}
+\usage{
+\method{plot}{prob_lnorm}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_norm}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (log normal)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_lnorm(meanlog = 0, sdlog = 1, lb = 0, ub = 1)
+plot(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_lnorm}} to calculate results
+
+\code{\link{plot.prob_lnorm}} to plot results
+}
diff --git a/radiant.basics/man/plot.prob_norm.Rd b/radiant.basics/man/plot.prob_norm.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..afe3f850a20c1294753cb48243b4f6dd1cf5e628
--- /dev/null
+++ b/radiant.basics/man/plot.prob_norm.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_norm}
+\alias{plot.prob_norm}
+\title{Plot method for the probability calculator (normal)}
+\usage{
+\method{plot}{prob_norm}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_norm}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (normal)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_norm(mean = 0, stdev = 1, ub = 0)
+plot(result)
+
+}
+\seealso{
+\code{\link{prob_norm}} to calculate results
+
+\code{\link{summary.prob_norm}} to summarize results
+}
diff --git a/radiant.basics/man/plot.prob_pois.Rd b/radiant.basics/man/plot.prob_pois.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..7cf291206eecdba6835687df5a4b5189b28038ce
--- /dev/null
+++ b/radiant.basics/man/plot.prob_pois.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_pois}
+\alias{plot.prob_pois}
+\title{Plot method for the probability calculator (poisson)}
+\usage{
+\method{plot}{prob_pois}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_pois}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (poisson)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_pois(lambda = 1, ub = 3)
+plot(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_pois}} to calculate results
+
+\code{\link{summary.prob_pois}} to summarize results
+}
diff --git a/radiant.basics/man/plot.prob_tdist.Rd b/radiant.basics/man/plot.prob_tdist.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..34ec0be99907549abd959e60a5ad92080ddd283d
--- /dev/null
+++ b/radiant.basics/man/plot.prob_tdist.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_tdist}
+\alias{plot.prob_tdist}
+\title{Plot method for the probability calculator (t-distribution)}
+\usage{
+\method{plot}{prob_tdist}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_tdist}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (t-distribution)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_tdist(df = 10, ub = 2.228)
+plot(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_tdist}} to calculate results
+
+\code{\link{summary.prob_tdist}} to summarize results
+}
diff --git a/radiant.basics/man/plot.prob_unif.Rd b/radiant.basics/man/plot.prob_unif.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..d5a763cd28dfb79eb00b542445fa8eca57b93c23
--- /dev/null
+++ b/radiant.basics/man/plot.prob_unif.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{plot.prob_unif}
+\alias{plot.prob_unif}
+\title{Plot method for the probability calculator (uniform)}
+\usage{
+\method{plot}{prob_unif}(x, type = "values", ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{prob_unif}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the probability calculator (uniform)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_unif(min = 0, max = 1, ub = 0.3)
+plot(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_unif}} to calculate results
+
+\code{\link{summary.prob_unif}} to summarize results
+}
diff --git a/radiant.basics/man/plot.single_mean.Rd b/radiant.basics/man/plot.single_mean.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..cf5d912ae1c75ebd1c6b69a5b1243d3c9a8c5877
--- /dev/null
+++ b/radiant.basics/man/plot.single_mean.Rd
@@ -0,0 +1,35 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/single_mean.R
+\name{plot.single_mean}
+\alias{plot.single_mean}
+\title{Plot method for the single_mean function}
+\usage{
+\method{plot}{single_mean}(x, plots = "hist", shiny = FALSE, custom = FALSE, ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{single_mean}}}
+
+\item{plots}{Plots to generate. "hist" shows a histogram of the data along with vertical lines that indicate the sample mean and the confidence interval. "simulate" shows the location of the sample mean and the comparison value (comp_value). Simulation is used to demonstrate the sampling variability in the data under the null-hypothesis}
+
+\item{shiny}{Did the function call originate inside a shiny app}
+
+\item{custom}{Logical (TRUE, FALSE) to indicate if ggplot object (or list of ggplot objects) should be returned. This option can be used to customize plots (e.g., add a title, change x and y labels, etc.). See examples and \url{https://ggplot2.tidyverse.org/} for options.}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the single_mean function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/single_mean.html} for an example in Radiant
+}
+\examples{
+result <- single_mean(diamonds, "price", comp_value = 3500)
+plot(result, plots = c("hist", "simulate"))
+
+}
+\seealso{
+\code{\link{single_mean}} to generate the result
+
+\code{\link{summary.single_mean}} to summarize results
+}
diff --git a/radiant.basics/man/plot.single_prop.Rd b/radiant.basics/man/plot.single_prop.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..6d63422c5490d9b60f7dc0f026e571735fe5b01b
--- /dev/null
+++ b/radiant.basics/man/plot.single_prop.Rd
@@ -0,0 +1,35 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/single_prop.R
+\name{plot.single_prop}
+\alias{plot.single_prop}
+\title{Plot method for the single_prop function}
+\usage{
+\method{plot}{single_prop}(x, plots = "bar", shiny = FALSE, custom = FALSE, ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{single_prop}}}
+
+\item{plots}{Plots to generate. "bar" shows a bar chart of the data. The "simulate" chart shows the location of the sample proportion and the comparison value (comp_value). Simulation is used to demonstrate the sampling variability in the data under the null-hypothesis}
+
+\item{shiny}{Did the function call originate inside a shiny app}
+
+\item{custom}{Logical (TRUE, FALSE) to indicate if ggplot object (or list of ggplot objects) should be returned. This option can be used to customize plots (e.g., add a title, change x and y labels, etc.). See examples and \url{https://ggplot2.tidyverse.org/} for options.}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Plot method for the single_prop function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/single_prop.html} for an example in Radiant
+}
+\examples{
+result <- single_prop(titanic, "survived", lev = "Yes", comp_value = 0.5, alternative = "less")
+plot(result, plots = c("bar", "simulate"))
+
+}
+\seealso{
+\code{\link{single_prop}} to generate the result
+
+\code{\link{summary.single_prop}} to summarize the results
+}
diff --git a/radiant.basics/man/print.rcorr.Rd b/radiant.basics/man/print.rcorr.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..06d15876bbb75409e525106fcc6e7759a44f9d60
--- /dev/null
+++ b/radiant.basics/man/print.rcorr.Rd
@@ -0,0 +1,16 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/correlation.R
+\name{print.rcorr}
+\alias{print.rcorr}
+\title{Print method for the correlation function}
+\usage{
+\method{print}{rcorr}(x, ...)
+}
+\arguments{
+\item{x}{Return value from \code{\link{correlation}}}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Print method for the correlation function
+}
diff --git a/radiant.basics/man/prob_binom.Rd b/radiant.basics/man/prob_binom.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..d49389aebbad1cec74e01b91435dcdbef571f7c5
--- /dev/null
+++ b/radiant.basics/man/prob_binom.Rd
@@ -0,0 +1,38 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_binom}
+\alias{prob_binom}
+\title{Probability calculator for the binomial distribution}
+\usage{
+prob_binom(n, p, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{n}{Number of trials}
+
+\item{p}{Probability}
+
+\item{lb}{Lower bound on the number of successes}
+
+\item{ub}{Upper bound on the number of successes}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for the binomial distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_binom(n = 10, p = 0.3, ub = 3)
+
+}
+\seealso{
+\code{\link{summary.prob_binom}} to summarize results
+
+\code{\link{plot.prob_binom}} to plot results
+}
diff --git a/radiant.basics/man/prob_chisq.Rd b/radiant.basics/man/prob_chisq.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..2accf21595abb8b61e55b97867abf7b1608e1caa
--- /dev/null
+++ b/radiant.basics/man/prob_chisq.Rd
@@ -0,0 +1,36 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_chisq}
+\alias{prob_chisq}
+\title{Probability calculator for the chi-squared distribution}
+\usage{
+prob_chisq(df, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{df}{Degrees of freedom}
+
+\item{lb}{Lower bound (default is 0)}
+
+\item{ub}{Upper bound (default is Inf)}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for the chi-squared distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_chisq(df = 1, ub = 3.841)
+
+}
+\seealso{
+\code{\link{summary.prob_chisq}} to summarize results
+
+\code{\link{plot.prob_chisq}} to plot results
+}
diff --git a/radiant.basics/man/prob_disc.Rd b/radiant.basics/man/prob_disc.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..11d4798a7b787d11fff170873c3223ff28f6f08c
--- /dev/null
+++ b/radiant.basics/man/prob_disc.Rd
@@ -0,0 +1,39 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_disc}
+\alias{prob_disc}
+\title{Probability calculator for a discrete distribution}
+\usage{
+prob_disc(v, p, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{v}{Values}
+
+\item{p}{Probabilities}
+
+\item{lb}{Lower bound on the number of successes}
+
+\item{ub}{Upper bound on the number of successes}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for a discrete distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_disc(v = 1:6, p = 1 / 6, pub = 0.95)
+prob_disc(v = 1:6, p = c(2 / 6, 2 / 6, 1 / 12, 1 / 12, 1 / 12, 1 / 12), pub = 0.95)
+
+}
+\seealso{
+\code{\link{summary.prob_disc}} to summarize results
+
+\code{\link{plot.prob_disc}} to plot results
+}
diff --git a/radiant.basics/man/prob_expo.Rd b/radiant.basics/man/prob_expo.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..5091759736ea481cc0c2c51495c962c46d5ac33b
--- /dev/null
+++ b/radiant.basics/man/prob_expo.Rd
@@ -0,0 +1,36 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_expo}
+\alias{prob_expo}
+\title{Probability calculator for the exponential distribution}
+\usage{
+prob_expo(rate, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{rate}{Rate}
+
+\item{lb}{Lower bound (default is 0)}
+
+\item{ub}{Upper bound (default is Inf)}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for the exponential distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_expo(rate = 1, ub = 2.996)
+
+}
+\seealso{
+\code{\link{summary.prob_expo}} to summarize results
+
+\code{\link{plot.prob_expo}} to plot results
+}
diff --git a/radiant.basics/man/prob_fdist.Rd b/radiant.basics/man/prob_fdist.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..aeac2d30c9f81944a775f2c02fa688fb545dd970
--- /dev/null
+++ b/radiant.basics/man/prob_fdist.Rd
@@ -0,0 +1,38 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_fdist}
+\alias{prob_fdist}
+\title{Probability calculator for the F-distribution}
+\usage{
+prob_fdist(df1, df2, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{df1}{Degrees of freedom}
+
+\item{df2}{Degrees of freedom}
+
+\item{lb}{Lower bound (default is 0)}
+
+\item{ub}{Upper bound (default is Inf)}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for the F-distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_fdist(df1 = 10, df2 = 10, ub = 2.978)
+
+}
+\seealso{
+\code{\link{summary.prob_fdist}} to summarize results
+
+\code{\link{plot.prob_fdist}} to plot results
+}
diff --git a/radiant.basics/man/prob_lnorm.Rd b/radiant.basics/man/prob_lnorm.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..74ab5373280b69dd23928f5b2990035e8e8c730d
--- /dev/null
+++ b/radiant.basics/man/prob_lnorm.Rd
@@ -0,0 +1,38 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_lnorm}
+\alias{prob_lnorm}
+\title{Probability calculator for the log normal distribution}
+\usage{
+prob_lnorm(meanlog, sdlog, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{meanlog}{Mean of the distribution on the log scale}
+
+\item{sdlog}{Standard deviation of the distribution on the log scale}
+
+\item{lb}{Lower bound (default is -Inf)}
+
+\item{ub}{Upper bound (default is Inf)}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for the log normal distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_lnorm(meanlog = 0, sdlog = 1, lb = 0, ub = 1)
+
+}
+\seealso{
+\code{\link{summary.prob_lnorm}} to summarize results
+
+\code{\link{plot.prob_lnorm}} to plot results
+}
diff --git a/radiant.basics/man/prob_norm.Rd b/radiant.basics/man/prob_norm.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..63193d14364958159fdd276ef671bcbf68ec319b
--- /dev/null
+++ b/radiant.basics/man/prob_norm.Rd
@@ -0,0 +1,38 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_norm}
+\alias{prob_norm}
+\title{Probability calculator for the normal distribution}
+\usage{
+prob_norm(mean, stdev, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{mean}{Mean}
+
+\item{stdev}{Standard deviation}
+
+\item{lb}{Lower bound (default is -Inf)}
+
+\item{ub}{Upper bound (default is Inf)}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for the normal distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_norm(mean = 0, stdev = 1, ub = 0)
+
+}
+\seealso{
+\code{\link{summary.prob_norm}} to summarize results
+
+\code{\link{plot.prob_norm}} to plot results
+}
diff --git a/radiant.basics/man/prob_pois.Rd b/radiant.basics/man/prob_pois.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..68c38162073edca98f2145e1455b403abf6bcb17
--- /dev/null
+++ b/radiant.basics/man/prob_pois.Rd
@@ -0,0 +1,36 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_pois}
+\alias{prob_pois}
+\title{Probability calculator for the poisson distribution}
+\usage{
+prob_pois(lambda, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{lambda}{Rate}
+
+\item{lb}{Lower bound (default is 0)}
+
+\item{ub}{Upper bound (default is Inf)}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for the poisson distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_pois(lambda = 1, ub = 3)
+
+}
+\seealso{
+\code{\link{summary.prob_pois}} to summarize results
+
+\code{\link{plot.prob_pois}} to plot results
+}
diff --git a/radiant.basics/man/prob_tdist.Rd b/radiant.basics/man/prob_tdist.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..7d108993b10a6e8b6366ef430b3fadbe3df3a116
--- /dev/null
+++ b/radiant.basics/man/prob_tdist.Rd
@@ -0,0 +1,36 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_tdist}
+\alias{prob_tdist}
+\title{Probability calculator for the t-distribution}
+\usage{
+prob_tdist(df, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{df}{Degrees of freedom}
+
+\item{lb}{Lower bound (default is -Inf)}
+
+\item{ub}{Upper bound (default is Inf)}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for the t-distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_tdist(df = 10, ub = 2.228)
+
+}
+\seealso{
+\code{\link{summary.prob_tdist}} to summarize results
+
+\code{\link{plot.prob_tdist}} to plot results
+}
diff --git a/radiant.basics/man/prob_unif.Rd b/radiant.basics/man/prob_unif.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..85ea27afa1ea6bbf6c11ef03d6290f200b7441d7
--- /dev/null
+++ b/radiant.basics/man/prob_unif.Rd
@@ -0,0 +1,38 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{prob_unif}
+\alias{prob_unif}
+\title{Probability calculator for the uniform distribution}
+\usage{
+prob_unif(min, max, lb = NA, ub = NA, plb = NA, pub = NA, dec = 3)
+}
+\arguments{
+\item{min}{Minimum value}
+
+\item{max}{Maximum value}
+
+\item{lb}{Lower bound (default = 0)}
+
+\item{ub}{Upper bound (default = 1)}
+
+\item{plb}{Lower probability bound}
+
+\item{pub}{Upper probability bound}
+
+\item{dec}{Number of decimals to show}
+}
+\description{
+Probability calculator for the uniform distribution
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+prob_unif(min = 0, max = 1, ub = 0.3)
+
+}
+\seealso{
+\code{\link{summary.prob_unif}} to summarize results
+
+\code{\link{plot.prob_unif}} to plot results
+}
diff --git a/radiant.basics/man/radiant.basics.Rd b/radiant.basics/man/radiant.basics.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..d9c36135096832917e619567ff45248403bf90d8
--- /dev/null
+++ b/radiant.basics/man/radiant.basics.Rd
@@ -0,0 +1,24 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/aaa.R, R/radiant.R
+\name{radiant.basics}
+\alias{radiant.basics}
+\title{radiant.basics}
+\usage{
+radiant.basics(state, ...)
+}
+\arguments{
+\item{state}{Path to state file to load}
+
+\item{...}{additional arguments to pass to shiny::runApp (e.g, port = 8080)}
+}
+\description{
+Launch radiant.basics in the default web browser
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/} for documentation and tutorials
+}
+\examples{
+\dontrun{
+radiant.basics()
+}
+}
diff --git a/radiant.basics/man/radiant.basics_viewer.Rd b/radiant.basics/man/radiant.basics_viewer.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..4a8aad4ed724bc9018fa773e264b5b7a13aacb1c
--- /dev/null
+++ b/radiant.basics/man/radiant.basics_viewer.Rd
@@ -0,0 +1,24 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/radiant.R
+\name{radiant.basics_viewer}
+\alias{radiant.basics_viewer}
+\title{Launch radiant.basics in the Rstudio viewer}
+\usage{
+radiant.basics_viewer(state, ...)
+}
+\arguments{
+\item{state}{Path to state file to load}
+
+\item{...}{additional arguments to pass to shiny::runApp (e.g, port = 8080)}
+}
+\description{
+Launch radiant.basics in the Rstudio viewer
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/} for documentation and tutorials
+}
+\examples{
+\dontrun{
+radiant.basics_viewer()
+}
+}
diff --git a/radiant.basics/man/radiant.basics_window.Rd b/radiant.basics/man/radiant.basics_window.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..7d58b514e6cbdd19a3756b3d702633863f2588ae
--- /dev/null
+++ b/radiant.basics/man/radiant.basics_window.Rd
@@ -0,0 +1,24 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/radiant.R
+\name{radiant.basics_window}
+\alias{radiant.basics_window}
+\title{Launch radiant.basics in an Rstudio window}
+\usage{
+radiant.basics_window(state, ...)
+}
+\arguments{
+\item{state}{Path to state file to load}
+
+\item{...}{additional arguments to pass to shiny::runApp (e.g, port = 8080)}
+}
+\description{
+Launch radiant.basics in an Rstudio window
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/} for documentation and tutorials
+}
+\examples{
+\dontrun{
+radiant.basics_window()
+}
+}
diff --git a/radiant.basics/man/salary.Rd b/radiant.basics/man/salary.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..ef7bdc1fc348a3f2b1f15a6abbc83794fb6c20e4
--- /dev/null
+++ b/radiant.basics/man/salary.Rd
@@ -0,0 +1,19 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/aaa.R
+\docType{data}
+\name{salary}
+\alias{salary}
+\title{Salaries for Professors}
+\format{
+A data frame with 397 rows and 6 variables
+}
+\usage{
+data(salary)
+}
+\description{
+Salaries for Professors
+}
+\details{
+2008-2009 nine-month salary for professors in a college in the US. Description provided in attr(salary,description")
+}
+\keyword{datasets}
diff --git a/radiant.basics/man/single_mean.Rd b/radiant.basics/man/single_mean.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..9fe0c8cf3de8a901b6198a3758a6e6de3709d7de
--- /dev/null
+++ b/radiant.basics/man/single_mean.Rd
@@ -0,0 +1,49 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/single_mean.R
+\name{single_mean}
+\alias{single_mean}
+\title{Compare a sample mean to a population mean}
+\usage{
+single_mean(
+ dataset,
+ var,
+ comp_value = 0,
+ alternative = "two.sided",
+ conf_lev = 0.95,
+ data_filter = "",
+ envir = parent.frame()
+)
+}
+\arguments{
+\item{dataset}{Dataset}
+
+\item{var}{The variable selected for the mean comparison}
+
+\item{comp_value}{Population value to compare to the sample mean}
+
+\item{alternative}{The alternative hypothesis ("two.sided", "greater", or "less")}
+
+\item{conf_lev}{Span for the confidence interval}
+
+\item{data_filter}{Expression entered in, e.g., Data > View to filter the dataset in Radiant. The expression should be a string (e.g., "price > 10000")}
+
+\item{envir}{Environment to extract data from}
+}
+\value{
+A list of variables defined in single_mean as an object of class single_mean
+}
+\description{
+Compare a sample mean to a population mean
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/single_mean.html} for an example in Radiant
+}
+\examples{
+single_mean(diamonds, "price") \%>\% str()
+
+}
+\seealso{
+\code{\link{summary.single_mean}} to summarize results
+
+\code{\link{plot.single_mean}} to plot results
+}
diff --git a/radiant.basics/man/single_prop.Rd b/radiant.basics/man/single_prop.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..3692d995597d6f6c467de6bdfbae4008f131b3f4
--- /dev/null
+++ b/radiant.basics/man/single_prop.Rd
@@ -0,0 +1,56 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/single_prop.R
+\name{single_prop}
+\alias{single_prop}
+\title{Compare a sample proportion to a population proportion}
+\usage{
+single_prop(
+ dataset,
+ var,
+ lev = "",
+ comp_value = 0.5,
+ alternative = "two.sided",
+ conf_lev = 0.95,
+ test = "binom",
+ data_filter = "",
+ envir = parent.frame()
+)
+}
+\arguments{
+\item{dataset}{Dataset}
+
+\item{var}{The variable selected for the proportion comparison}
+
+\item{lev}{The factor level selected for the proportion comparison}
+
+\item{comp_value}{Population value to compare to the sample proportion}
+
+\item{alternative}{The alternative hypothesis ("two.sided", "greater", or "less")}
+
+\item{conf_lev}{Span of the confidence interval}
+
+\item{test}{bionomial exact test ("binom") or Z-test ("z")}
+
+\item{data_filter}{Expression entered in, e.g., Data > View to filter the dataset in Radiant. The expression should be a string (e.g., "price > 10000")}
+
+\item{envir}{Environment to extract data from}
+}
+\value{
+A list of variables used in single_prop as an object of class single_prop
+}
+\description{
+Compare a sample proportion to a population proportion
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/single_prop.html} for an example in Radiant
+}
+\examples{
+single_prop(titanic, "survived") \%>\% str()
+single_prop(titanic, "survived", lev = "Yes", comp_value = 0.5, alternative = "less") \%>\% str()
+
+}
+\seealso{
+\code{\link{summary.single_prop}} to summarize the results
+
+\code{\link{plot.single_prop}} to plot the results
+}
diff --git a/radiant.basics/man/summary.compare_means.Rd b/radiant.basics/man/summary.compare_means.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..96aafc43879e6c8c82819aed7c8117ab565cc6a9
--- /dev/null
+++ b/radiant.basics/man/summary.compare_means.Rd
@@ -0,0 +1,33 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/compare_means.R
+\name{summary.compare_means}
+\alias{summary.compare_means}
+\title{Summary method for the compare_means function}
+\usage{
+\method{summary}{compare_means}(object, show = FALSE, dec = 3, ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{compare_means}}}
+
+\item{show}{Show additional output (i.e., t.value, df, and confidence interval)}
+
+\item{dec}{Number of decimals to show}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the compare_means function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/compare_means.html} for an example in Radiant
+}
+\examples{
+result <- compare_means(diamonds, "cut", "price")
+summary(result)
+
+}
+\seealso{
+\code{\link{compare_means}} to calculate results
+
+\code{\link{plot.compare_means}} to plot results
+}
diff --git a/radiant.basics/man/summary.compare_props.Rd b/radiant.basics/man/summary.compare_props.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..5d028da495a8c7a5057952c2a0713f7913818f45
--- /dev/null
+++ b/radiant.basics/man/summary.compare_props.Rd
@@ -0,0 +1,33 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/compare_props.R
+\name{summary.compare_props}
+\alias{summary.compare_props}
+\title{Summary method for the compare_props function}
+\usage{
+\method{summary}{compare_props}(object, show = FALSE, dec = 3, ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{compare_props}}}
+
+\item{show}{Show additional output (i.e., chisq.value, df, and confidence interval)}
+
+\item{dec}{Number of decimals to show}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the compare_props function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/compare_props.html} for an example in Radiant
+}
+\examples{
+result <- compare_props(titanic, "pclass", "survived")
+summary(result)
+
+}
+\seealso{
+\code{\link{compare_props}} to calculate results
+
+\code{\link{plot.compare_props}} to plot results
+}
diff --git a/radiant.basics/man/summary.correlation.Rd b/radiant.basics/man/summary.correlation.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..0c6a824525005fcc1ff79d2a2b837834c3bc89f6
--- /dev/null
+++ b/radiant.basics/man/summary.correlation.Rd
@@ -0,0 +1,35 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/correlation.R
+\name{summary.correlation}
+\alias{summary.correlation}
+\title{Summary method for the correlation function}
+\usage{
+\method{summary}{correlation}(object, cutoff = 0, covar = FALSE, dec = 2, ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{correlation}}}
+
+\item{cutoff}{Show only correlations larger than the cutoff in absolute value. Default is a cutoff of 0}
+
+\item{covar}{Show the covariance matrix (default is FALSE)}
+
+\item{dec}{Number of decimals to show}
+
+\item{...}{further arguments passed to or from other methods.}
+}
+\description{
+Summary method for the correlation function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/correlation.html} for an example in Radiant
+}
+\examples{
+result <- correlation(diamonds, c("price", "carat", "table"))
+summary(result, cutoff = .3)
+
+}
+\seealso{
+\code{\link{correlation}} to calculate results
+
+\code{\link{plot.correlation}} to plot results
+}
diff --git a/radiant.basics/man/summary.cross_tabs.Rd b/radiant.basics/man/summary.cross_tabs.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..4f2e0207b591899486f15cd5ebd21a98db0ffa5c
--- /dev/null
+++ b/radiant.basics/man/summary.cross_tabs.Rd
@@ -0,0 +1,33 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/cross_tabs.R
+\name{summary.cross_tabs}
+\alias{summary.cross_tabs}
+\title{Summary method for the cross_tabs function}
+\usage{
+\method{summary}{cross_tabs}(object, check = "", dec = 2, ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{cross_tabs}}}
+
+\item{check}{Show table(s) for variables var1 and var2. "observed" for the observed frequencies table, "expected" for the expected frequencies table (i.e., frequencies that would be expected if the null hypothesis holds), "chi_sq" for the contribution to the overall chi-squared statistic for each cell (i.e., (o - e)^2 / e), "dev_std" for the standardized differences between the observed and expected frequencies (i.e., (o - e) / sqrt(e)), and "dev_perc" for the percentage difference between the observed and expected frequencies (i.e., (o - e) / e)}
+
+\item{dec}{Number of decimals to show}
+
+\item{...}{further arguments passed to or from other methods.}
+}
+\description{
+Summary method for the cross_tabs function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/cross_tabs.html} for an example in Radiant
+}
+\examples{
+result <- cross_tabs(newspaper, "Income", "Newspaper")
+summary(result, check = c("observed", "expected", "chi_sq"))
+
+}
+\seealso{
+\code{\link{cross_tabs}} to calculate results
+
+\code{\link{plot.cross_tabs}} to plot results
+}
diff --git a/radiant.basics/man/summary.goodness.Rd b/radiant.basics/man/summary.goodness.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..2163d863beb0ff5c1f1212cccf78637142a20526
--- /dev/null
+++ b/radiant.basics/man/summary.goodness.Rd
@@ -0,0 +1,34 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/goodness.R
+\name{summary.goodness}
+\alias{summary.goodness}
+\title{Summary method for the goodness function}
+\usage{
+\method{summary}{goodness}(object, check = "", dec = 2, ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{goodness}}}
+
+\item{check}{Show table(s) for the selected variable (var). "observed" for the observed frequencies table, "expected" for the expected frequencies table (i.e., frequencies that would be expected if the null hypothesis holds), "chi_sq" for the contribution to the overall chi-squared statistic for each cell (i.e., (o - e)^2 / e), "dev_std" for the standardized differences between the observed and expected frequencies (i.e., (o - e) / sqrt(e)), and "dev_perc" for the percentage difference between the observed and expected frequencies (i.e., (o - e) / e)}
+
+\item{dec}{Number of decimals to show}
+
+\item{...}{further arguments passed to or from other methods.}
+}
+\description{
+Summary method for the goodness function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/goodness} for an example in Radiant
+}
+\examples{
+result <- goodness(newspaper, "Income", c(.3, .7))
+summary(result, check = c("observed", "expected", "chi_sq"))
+goodness(newspaper, "Income", c(1 / 3, 2 / 3)) \%>\% summary("observed")
+
+}
+\seealso{
+\code{\link{goodness}} to calculate results
+
+\code{\link{plot.goodness}} to plot results
+}
diff --git a/radiant.basics/man/summary.prob_binom.Rd b/radiant.basics/man/summary.prob_binom.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..2be5878e633abc64f5c90f6c30ced937ed5059ea
--- /dev/null
+++ b/radiant.basics/man/summary.prob_binom.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_binom}
+\alias{summary.prob_binom}
+\title{Summary method for the probability calculator (binomial)}
+\usage{
+\method{summary}{prob_binom}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_binom}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (binomial)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_binom(n = 10, p = 0.3, ub = 3)
+summary(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_binom}} to calculate results
+
+\code{\link{plot.prob_binom}} to plot results
+}
diff --git a/radiant.basics/man/summary.prob_chisq.Rd b/radiant.basics/man/summary.prob_chisq.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..dea05f3902f14fc8bebf34ed5ef97200a5014987
--- /dev/null
+++ b/radiant.basics/man/summary.prob_chisq.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_chisq}
+\alias{summary.prob_chisq}
+\title{Summary method for the probability calculator (Chi-squared distribution)}
+\usage{
+\method{summary}{prob_chisq}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_chisq}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (Chi-squared distribution)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_chisq(df = 1, ub = 3.841)
+summary(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_chisq}} to calculate results
+
+\code{\link{plot.prob_chisq}} to plot results
+}
diff --git a/radiant.basics/man/summary.prob_disc.Rd b/radiant.basics/man/summary.prob_disc.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..4dc69d30b1cbb9136f822e1151a26b48603e6a57
--- /dev/null
+++ b/radiant.basics/man/summary.prob_disc.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_disc}
+\alias{summary.prob_disc}
+\title{Summary method for the probability calculator (discrete)}
+\usage{
+\method{summary}{prob_disc}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_disc}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (discrete)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_disc(v = 1:6, p = c(2 / 6, 2 / 6, 1 / 12, 1 / 12, 1 / 12, 1 / 12), pub = 0.95)
+summary(result, type = "probs")
+
+}
+\seealso{
+\code{\link{prob_disc}} to calculate results
+
+\code{\link{plot.prob_disc}} to plot results
+}
diff --git a/radiant.basics/man/summary.prob_expo.Rd b/radiant.basics/man/summary.prob_expo.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..9a526abf1c3e4fcce18cf2c115e3ea9b2d6b3a1b
--- /dev/null
+++ b/radiant.basics/man/summary.prob_expo.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_expo}
+\alias{summary.prob_expo}
+\title{Summary method for the probability calculator (exponential)}
+\usage{
+\method{summary}{prob_expo}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_expo}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (exponential)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_expo(rate = 1, ub = 2.996)
+summary(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_expo}} to calculate results
+
+\code{\link{plot.prob_expo}} to plot results
+}
diff --git a/radiant.basics/man/summary.prob_fdist.Rd b/radiant.basics/man/summary.prob_fdist.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..d6a78dcef28ff0317a05790093c17706c973cbf1
--- /dev/null
+++ b/radiant.basics/man/summary.prob_fdist.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_fdist}
+\alias{summary.prob_fdist}
+\title{Summary method for the probability calculator (F-distribution)}
+\usage{
+\method{summary}{prob_fdist}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_fdist}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (F-distribution)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_fdist(df1 = 10, df2 = 10, ub = 2.978)
+summary(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_fdist}} to calculate results
+
+\code{\link{plot.prob_fdist}} to plot results
+}
diff --git a/radiant.basics/man/summary.prob_lnorm.Rd b/radiant.basics/man/summary.prob_lnorm.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..a1348794597bf68acd0e5d671cebd7c76058629c
--- /dev/null
+++ b/radiant.basics/man/summary.prob_lnorm.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_lnorm}
+\alias{summary.prob_lnorm}
+\title{Summary method for the probability calculator (log normal)}
+\usage{
+\method{summary}{prob_lnorm}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_norm}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (log normal)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_lnorm(meanlog = 0, sdlog = 1, lb = 0, ub = 1)
+summary(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_lnorm}} to calculate results
+
+\code{\link{plot.prob_lnorm}} to summarize results
+}
diff --git a/radiant.basics/man/summary.prob_norm.Rd b/radiant.basics/man/summary.prob_norm.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..c90a47643574074af751ee02510ab9b9ed4f0452
--- /dev/null
+++ b/radiant.basics/man/summary.prob_norm.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_norm}
+\alias{summary.prob_norm}
+\title{Summary method for the probability calculator (normal)}
+\usage{
+\method{summary}{prob_norm}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_norm}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (normal)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_norm(mean = 0, stdev = 1, ub = 0)
+summary(result)
+
+}
+\seealso{
+\code{\link{prob_norm}} to calculate results
+
+\code{\link{plot.prob_norm}} to plot results
+}
diff --git a/radiant.basics/man/summary.prob_pois.Rd b/radiant.basics/man/summary.prob_pois.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..b61780c463e747bc825f0509e6a41d29c76a6fc7
--- /dev/null
+++ b/radiant.basics/man/summary.prob_pois.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_pois}
+\alias{summary.prob_pois}
+\title{Summary method for the probability calculator (poisson)}
+\usage{
+\method{summary}{prob_pois}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_pois}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (poisson)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_pois(lambda = 1, ub = 3)
+summary(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_pois}} to calculate results
+
+\code{\link{plot.prob_pois}} to plot results
+}
diff --git a/radiant.basics/man/summary.prob_tdist.Rd b/radiant.basics/man/summary.prob_tdist.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..41d85ab4c74d72ff838a49dc6eaf82b73be449cc
--- /dev/null
+++ b/radiant.basics/man/summary.prob_tdist.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_tdist}
+\alias{summary.prob_tdist}
+\title{Summary method for the probability calculator (t-distribution)}
+\usage{
+\method{summary}{prob_tdist}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_tdist}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (t-distribution)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_tdist(df = 10, ub = 2.228)
+summary(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_tdist}} to calculate results
+
+\code{\link{plot.prob_tdist}} to plot results
+}
diff --git a/radiant.basics/man/summary.prob_unif.Rd b/radiant.basics/man/summary.prob_unif.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..7364b92ce1b66367730b18ee99617a6b65bf1336
--- /dev/null
+++ b/radiant.basics/man/summary.prob_unif.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/prob_calc.R
+\name{summary.prob_unif}
+\alias{summary.prob_unif}
+\title{Summary method for the probability calculator (uniform)}
+\usage{
+\method{summary}{prob_unif}(object, type = "values", ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{prob_unif}}}
+
+\item{type}{Probabilities ("probs") or values ("values")}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the probability calculator (uniform)
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/prob_calc.html} for an example in Radiant
+}
+\examples{
+result <- prob_unif(min = 0, max = 1, ub = 0.3)
+summary(result, type = "values")
+
+}
+\seealso{
+\code{\link{prob_unif}} to calculate results
+
+\code{\link{plot.prob_unif}} to plot results
+}
diff --git a/radiant.basics/man/summary.single_mean.Rd b/radiant.basics/man/summary.single_mean.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..7d06b9cbf49a064d0c0c6b5fb2548d11f42bfff4
--- /dev/null
+++ b/radiant.basics/man/summary.single_mean.Rd
@@ -0,0 +1,34 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/single_mean.R
+\name{summary.single_mean}
+\alias{summary.single_mean}
+\title{Summary method for the single_mean function}
+\usage{
+\method{summary}{single_mean}(object, dec = 3, ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{single_mean}}}
+
+\item{dec}{Number of decimals to show}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the single_mean function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/single_mean.html} for an example in Radiant
+}
+\examples{
+result <- single_mean(diamonds, "price")
+summary(result)
+diamonds \%>\%
+ single_mean("price") \%>\%
+ summary()
+
+}
+\seealso{
+\code{\link{single_mean}} to generate the results
+
+\code{\link{plot.single_mean}} to plot results
+}
diff --git a/radiant.basics/man/summary.single_prop.Rd b/radiant.basics/man/summary.single_prop.Rd
new file mode 100644
index 0000000000000000000000000000000000000000..e683bfa294a9f1f807e661420fd54ffb7c319b9c
--- /dev/null
+++ b/radiant.basics/man/summary.single_prop.Rd
@@ -0,0 +1,31 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/single_prop.R
+\name{summary.single_prop}
+\alias{summary.single_prop}
+\title{Summary method for the single_prop function}
+\usage{
+\method{summary}{single_prop}(object, dec = 3, ...)
+}
+\arguments{
+\item{object}{Return value from \code{\link{single_prop}}}
+
+\item{dec}{Number of decimals to show}
+
+\item{...}{further arguments passed to or from other methods}
+}
+\description{
+Summary method for the single_prop function
+}
+\details{
+See \url{https://radiant-rstats.github.io/docs/basics/single_prop.html} for an example in Radiant
+}
+\examples{
+result <- single_prop(titanic, "survived", lev = "Yes", comp_value = 0.5, alternative = "less")
+summary(result)
+
+}
+\seealso{
+\code{\link{single_prop}} to generate the results
+
+\code{\link{plot.single_prop}} to plot the results
+}
diff --git a/radiant.basics/tests/testthat.R b/radiant.basics/tests/testthat.R
new file mode 100644
index 0000000000000000000000000000000000000000..5e9232f5f2e637e97d41b03bb04a29398117dca4
--- /dev/null
+++ b/radiant.basics/tests/testthat.R
@@ -0,0 +1,5 @@
+## use shift-cmd-t to run all tests
+library(testthat)
+test_check("radiant.basics")
+# if (interactive() && !exists("coverage_test")) devtools::run_examples()
+# devtools::run_examples(start = "single_prop")
diff --git a/radiant.basics/tests/testthat/test_stats.R b/radiant.basics/tests/testthat/test_stats.R
new file mode 100644
index 0000000000000000000000000000000000000000..165b986e716db9de6e20c5c3a72e1a242505b15f
--- /dev/null
+++ b/radiant.basics/tests/testthat/test_stats.R
@@ -0,0 +1,82 @@
+# library(radiant.basics)
+# library(testthat)
+
+trim <- function(x) gsub("^\\s+|\\s+$", "", x)
+
+context("Compare means")
+
+test_that("compare_means 1", {
+ result <- compare_means(diamonds, "cut", "price")
+ res1 <- capture.output(summary(result))[9] %>% trim()
+ # cat(paste0(res1, "\n"))
+ res2 <- "Fair 4,505.238 101 0 3,749.540 373.093 740.206"
+ expect_equal(res1, res2)
+ res1 <- capture.output(summary(result))[16] %>% trim()
+ # cat(paste0(res1, "\n"))
+ res2 <- "Fair = Good Fair not equal to Good 374.805 0.391"
+ expect_equal(res1, res2)
+})
+
+test_that("compare_means 2", {
+ result <- compare_means(diamonds, "cut", "price")
+ res1 <- capture_output(summary(result, show = TRUE))
+ # dput(res1)
+ res2 <- "Pairwise mean comparisons (t-test)\nData : diamonds \nVariables : cut, price \nSamples : independent \nConfidence: 0.95 \nAdjustment: None \n\n cut mean n n_missing sd se me\n Fair 4,505.238 101 0 3,749.540 373.093 740.206\n Good 4,130.433 275 0 3,730.354 224.949 442.848\n Very Good 3,959.916 677 0 3,895.899 149.732 293.995\n Premium 4,369.409 771 0 4,236.977 152.591 299.544\n Ideal 3,470.224 1,176 0 3,827.423 111.610 218.977\n\n Null hyp. Alt. hyp. diff p.value\n Fair = Good Fair not equal to Good 374.805 0.391 \n Fair = Very Good Fair not equal to Very Good 545.322 0.177 \n Fair = Premium Fair not equal to Premium 135.829 0.737 \n Fair = Ideal Fair not equal to Ideal 1035.014 0.009 \n Good = Very Good Good not equal to Very Good 170.517 0.528 \n Good = Premium Good not equal to Premium -238.976 0.38 \n Good = Ideal Good not equal to Ideal 660.209 0.009 \n Very Good = Premium Very Good not equal to Premium -409.493 0.056 \n Very Good = Ideal Very Good not equal to Ideal 489.692 0.009 \n Premium = Ideal Premium not equal to Ideal 899.185 < .001 \n se t.value df 2.5% 97.5% \n 435.661 0.860 177.365 -484.941 1234.551 \n 402.018 1.356 134.291 -249.783 1340.427 \n 403.091 0.337 135.759 -661.321 932.979 \n 389.429 2.658 118.618 263.879 1806.149 ** \n 270.225 0.631 528.529 -360.330 701.364 \n 271.820 -0.879 543.242 -772.922 294.971 \n 251.115 2.629 419.577 166.609 1153.809 ** \n 213.784 -1.915 1442.922 -828.853 9.868 . \n 186.752 2.622 1389.163 123.346 856.039 ** \n 189.052 4.756 1527.729 528.355 1270.015 ***\n\nSignif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1"
+ expect_equal(res1, res2)
+})
+
+context("Compare proportions")
+
+test_that("compare_props 1", {
+ result <- compare_props(titanic, "pclass", "survived")
+ res1 <- capture.output(summary(result))[9] %>% trim()
+ # cat(paste0(res1, "\n"))
+ res2 <- "1st 179 103 0.635 282 0 0.481 0.029 0.056"
+ expect_equal(res1, res2)
+})
+
+test_that("compare_props 2", {
+ result <- compare_props(titanic, "pclass", "survived")
+ res1 <- capture_output(summary(result, show = TRUE))
+ # dput(res1)
+ res2 <- "Pairwise proportion comparisons\nData : titanic \nVariables : pclass, survived \nLevel : in survived \nConfidence: 0.95 \nAdjustment: None \n\n pclass Yes No p n n_missing sd se me\n 1st 179 103 0.635 282 0 0.481 0.029 0.056\n 2nd 115 146 0.441 261 0 0.496 0.031 0.060\n 3rd 131 369 0.262 500 0 0.440 0.020 0.039\n\n Null hyp. Alt. hyp. diff p.value chisq.value df 2.5% 97.5%\n 1st = 2nd 1st not equal to 2nd 0.194 < .001 20.576 1 0.112 0.277\n 1st = 3rd 1st not equal to 3rd 0.373 < .001 104.704 1 0.305 0.441\n 2nd = 3rd 2nd not equal to 3rd 0.179 < .001 25.008 1 0.107 0.250\n \n ***\n ***\n ***\n\nSignif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1"
+ expect_equal(res1, res2)
+})
+
+context("Single proportion")
+
+test_that("single_prop 1", {
+ result <- single_prop(diamonds, "color")
+ expect_equal(result$lev, "D")
+ res1 <- capture_output(summary(result))
+ # dput(res1)
+ res2 <- "Single proportion test (binomial exact)\nData : diamonds \nVariable : color \nLevel : D in color \nConfidence: 0.95 \nNull hyp. : the proportion of D in color = 0.5 \nAlt. hyp. : the proportion of D in color not equal to 0.5 \n\n p ns n n_missing sd se me\n 0.127 382 3,000 0 0.333 0.006 0.012\n\n diff ns p.value 2.5% 97.5% \n -0.373 382 < .001 0.116 0.140 ***\n\nSignif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1"
+ expect_equal(res1, res2)
+})
+
+test_that("single_prop 2", {
+ result <- single_prop(diamonds, "clarity", lev = "IF", comp_value = 0.05)
+ expect_equal(result$lev, "IF")
+ res1 <- capture_output(summary(result))
+ # dput(res1)
+ res2 <- "Single proportion test (binomial exact)\nData : diamonds \nVariable : clarity \nLevel : IF in clarity \nConfidence: 0.95 \nNull hyp. : the proportion of IF in clarity = 0.05 \nAlt. hyp. : the proportion of IF in clarity not equal to 0.05 \n\n p ns n n_missing sd se me\n 0.033 99 3,000 0 0.179 0.003 0.006\n\n diff ns p.value 2.5% 97.5% \n -0.017 99 < .001 0.027 0.040 ***\n\nSignif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1"
+ expect_equal(res1, res2)
+})
+
+context("Single mean")
+
+test_that("single_mean 1", {
+ result <- single_mean(diamonds, "carat")
+ res1 <- capture.output(summary(result))[12] %>% trim()
+ # cat(paste0(res1, "\n"))
+ res2 <- "0.794 0.009 91.816 < .001 2999 0.777 0.811 ***"
+ expect_equal(res1, res2)
+})
+
+test_that("single_mean 2", {
+ result <- single_mean(titanic, "age", comp_value = 40)
+ res1 <- capture.output(summary(result))[12] %>% trim()
+ # cat(paste0(res1, "\n"))
+ res2 <- "-10.187 0.445 -22.9 < .001 1042 28.94 30.686 ***"
+ expect_equal(res1, res2)
+})
diff --git a/radiant.basics/vignettes/pkgdown/_clt.Rmd b/radiant.basics/vignettes/pkgdown/_clt.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..1d807bac2973a097bf881133c6fbf103d28c86b1
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/_clt.Rmd
@@ -0,0 +1,19 @@
+> Using random sampling to illustrate the Central Limit Theorem
+
+### What is the Central Limit Theorem?
+
+"In probability theory, the central limit theorem (CLT) states that, given certain conditions, the arithmetic mean of a sufficiently large number of iterates of independent random variables, each with a well-defined expected value and well-defined variance, will be approximately normally distributed, regardless of the underlying distribution. That is, suppose that a sample is obtained containing a large number of observations, each observation being randomly generated in a way that does not depend on the values of the other observations, and that the arithmetic average of the observed values is computed. If this procedure is performed many times, the central limit theorem says that the computed values of the average will be distributed according to the normal distribution (commonly known as a 'bell curve')."
+
+Source: Wikipedia
+
+## Sample
+
+To generate samples select a distribution from the `Distribution` dropdown and accept (or change) the default values. Then press `Sample` or press `CTRL-enter` (`CMD-enter` on mac) to run the simulation and show plots of the simulated data.
+
+### Khan on the CLT
+
+
+
+### R-functions
+
+For an overview of related R-functions used by Radiant for probability calculations see _Basics > Probability_
diff --git a/radiant.basics/vignettes/pkgdown/_compare_means.Rmd b/radiant.basics/vignettes/pkgdown/_compare_means.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..3dbe5793d24df6379ceb08b0bb3cfe15ff2fdfbe
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/_compare_means.Rmd
@@ -0,0 +1,120 @@
+> Compare the means of two or more variables or groups in the data
+
+The compare means t-test is used to compare the mean of a variable in one group to the mean of the same variable in one, or more, other groups. The null hypothesis for the difference between the groups in the population is set to zero. We test this hypothesis using sample data.
+
+We can perform either a one-tailed test (i.e., `less than` or `greater than`) or a two-tailed test (see the 'Alternative hypothesis' dropdown). We use one-tailed tests to evaluate if the available data provide evidence that the difference in sample means between groups is less than (or greater than ) zero.
+
+### Example: Professor salaries
+
+We have access to the nine-month academic salary for Assistant Professors, Associate Professors and Professors in a college in the U.S (2008-09). The data were collected as part of an on-going effort by the college's administration to monitor salary differences between male and female faculty members. The data has 397 observations and the following 6 variables.
+
+- rank = a factor with levels AsstProf, AssocProf, and Prof
+- discipline = a factor with levels A ("theoretical" departments) or B ("applied" departments)
+- yrs.since.phd = years since PhD
+- yrs.service = years of service
+- sex = a factor with levels Female and Male
+- salary = nine-month salary, in dollars
+
+The data are part of the CAR package and are linked to the book: Fox J. and Weisberg, S. (2011) An R Companion to Applied Regression, Second Edition Sage.
+
+Suppose we want to test if professors of lower rank earn lower salaries compared to those of higher rank. To test this hypothesis we first select professor `rank` and select `salary` as the numerical variable to compare across ranks. In the `Choose combinations` box select all available entries to conduct pair-wise comparisons across the three levels. Note that removing all entries will automatically select all combinations. We are interested in a one-sided hypothesis (i.e., `less than`).
+
+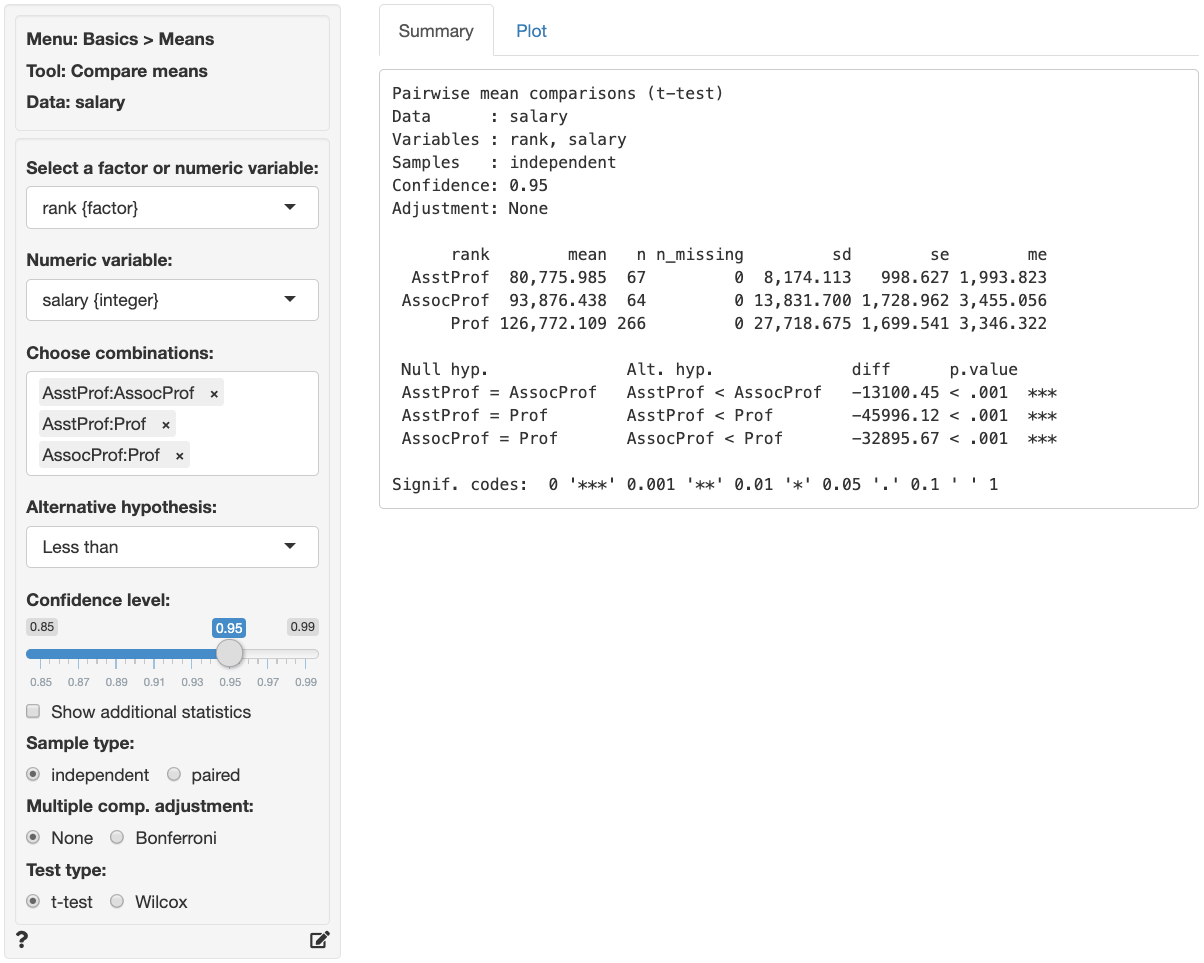
+Pairwise mean comparisons (t-test) +Data : salary +Variables : rank, salary +Samples : independent +Confidence: 0.95 +Adjustment: None + + rank mean n n_missing sd se me + AsstProf 80,775.985 67 0 8,174.113 998.627 1,993.823 + AssocProf 93,876.438 64 0 13,831.700 1,728.962 3,455.056 + Prof 126,772.109 266 0 27,718.675 1,699.541 3,346.322 + + Null hyp. Alt. hyp. diff p.value se t.value df 0% 95% + AsstProf = AssocProf AsstProf < AssocProf -13100.45 < .001 1996.639 -6.561 101.286 -Inf -9785.958 *** + AsstProf = Prof AsstProf < Prof -45996.12 < .001 1971.217 -23.334 324.340 -Inf -42744.474 *** + AssocProf = Prof AssocProf < Prof -32895.67 < .001 2424.407 -13.569 199.325 -Inf -28889.256 *** + +Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ++ +* `se` is the standard error (i.e., the standard deviation of the sampling distribution of `diff`) +* `t.value` is the _t_ statistic associated with `diff` that we can compare to a t-distribution (i.e., `diff` / `se`) +* `df` is the degrees of freedom associated with the statistical test. Note that the Welch approximation is used for the degrees of freedom +* `0% 95%` show the 95% confidence interval around the difference in sample means. These numbers provide a range within which the true population difference is likely to fall + +### Testing + +There are three approaches we can use to evaluate the null hypothesis. We will choose a significance level of 0.05.1 Of course, each approach will lead to the same conclusion. + +#### p.value + +Because each of the p.values is **smaller** than the significance level we reject the null hypothesis for each evaluated pair of professor ranks. The data suggest that associate professors make more than assistant professors and professors make more than assistant and associate professors. Note also the '***' that are used as an indicator for significance. + +#### confidence interval + +Because zero is **not** contained in any of the confidence intervals we reject the null hypothesis for each evaluated combination of ranks. Because our alternative hypothesis is `Less than` the confidence interval is actually an upper bound for the difference in salaries in the population at a 95% confidence level (i.e., -9785.958, -42744.474, and -28889.256) + +#### t.value + +Because the calculated t.values (-6.561, -23.334, and -13.569) are **smaller** than the corresponding _critical_ t.value we reject the null hypothesis for each evaluated combination of ranks. We can obtain the critical t.value by using the probability calculator in the _Basics_ menu. Using the test for assistant versus associate professors as an example, we find that for a t-distribution with 101.286 degrees of freedom (see `df`) the critical t.value is 1.66. We choose 0.05 as the lower probability bound because the alternative hypothesis is `Less than`. + +
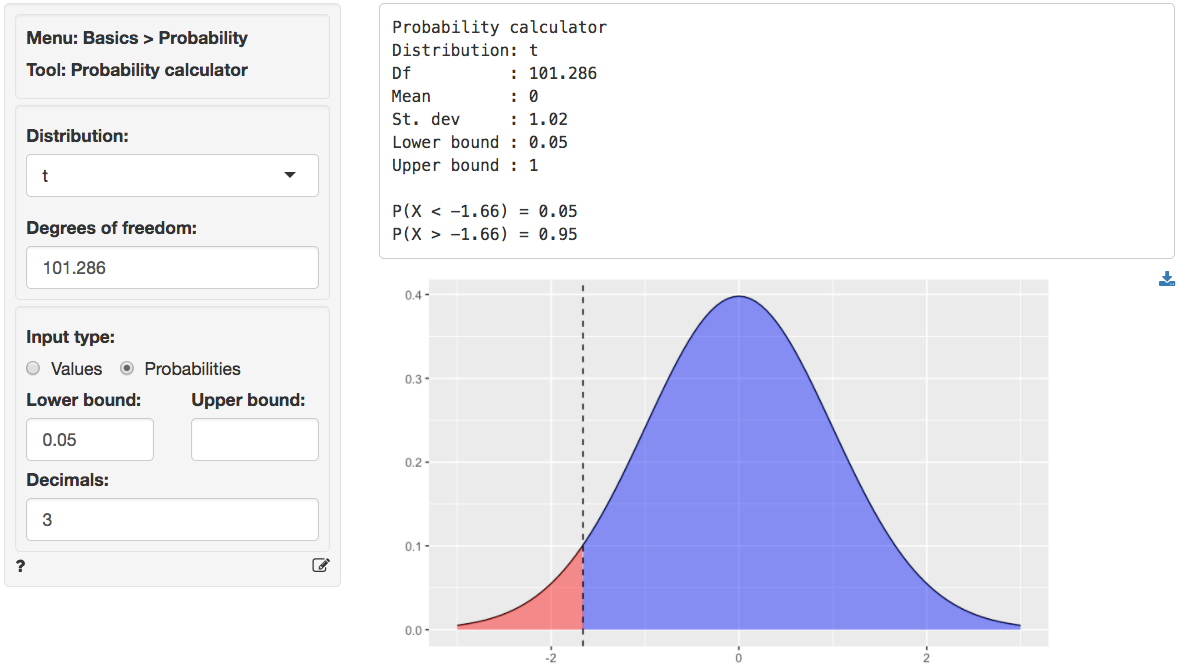
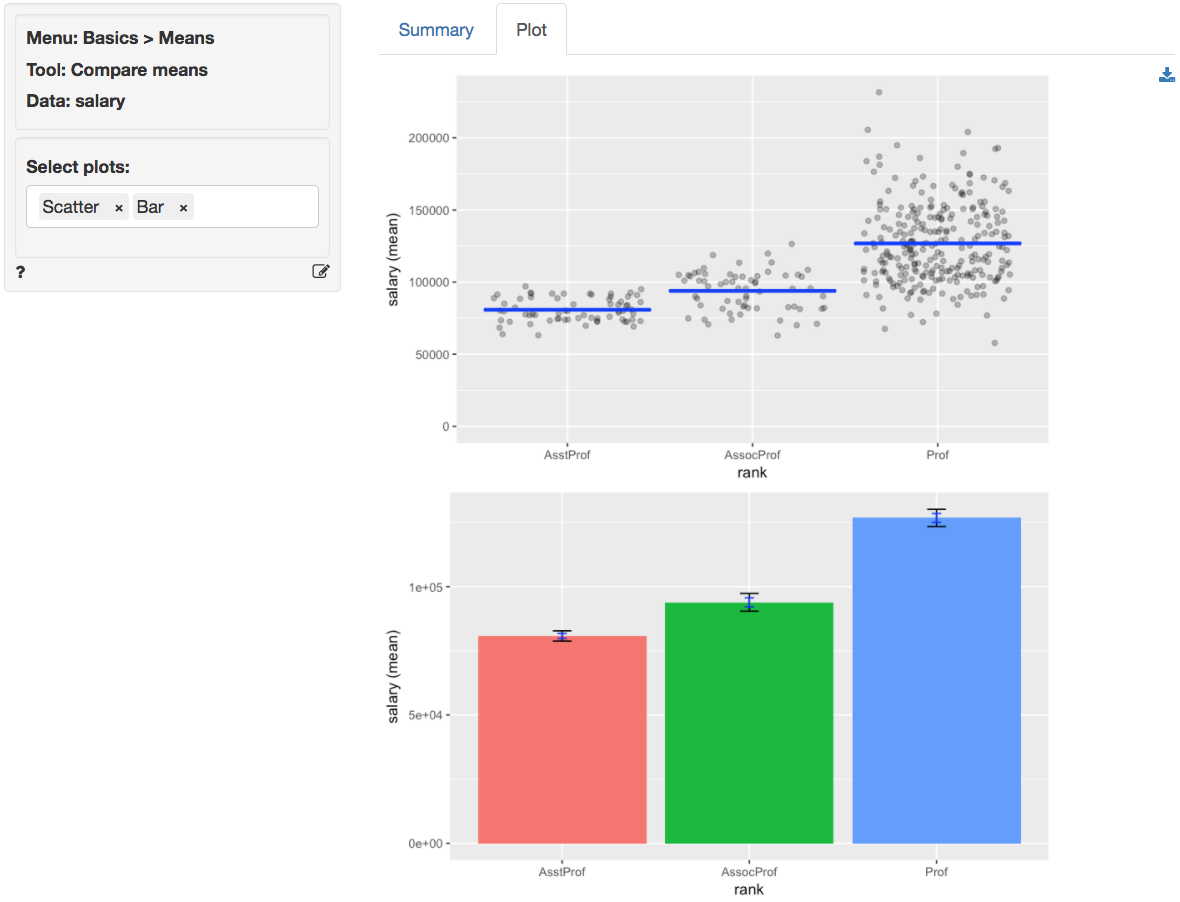
usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+Compare Means Hypothesis Test
+
+* This video shows how to conduct a compare means hypothesis test
+* Topics List:
+ - Calculate summary statistics by groups
+ - Setup a hypothesis test for compare means in Radiant
+ - Use the p.value and confidence interval to evaluate the hypothesis test
diff --git a/radiant.basics/vignettes/pkgdown/_compare_props.Rmd b/radiant.basics/vignettes/pkgdown/_compare_props.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..e56ad244c2942be564453bf30bf27a9f154ef74f
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/_compare_props.Rmd
@@ -0,0 +1,120 @@
+> Compare proportions for two or more groups in the data
+
+The compare proportions test is used to evaluate if the frequency of occurrence of some event, behavior, intention, etc. differs across groups. The null hypothesis for the difference in proportions across groups in the population is set to zero. We test this hypothesis using sample data.
+
+We can perform either a one-tailed test (i.e., `less than` or `greater than`) or a two-tailed test (see the `Alternative hypothesis` dropdown). A one-tailed test is useful if we want to evaluate if the available sample data suggest that, for example, the proportion of dropped calls is larger (or smaller) for one wireless provider compared to others.
+
+### Example
+
+We will use a sample from a dataset that describes the survival status of individual passengers on the Titanic. The principal source for data about Titanic passengers is the Encyclopedia Titanic. One of the original sources is Eaton & Haas (1994) Titanic: Triumph and Tragedy, Patrick Stephens Ltd, which includes a passenger list created by many researchers and edited by Michael A. Findlay. Lets focus on two variables in the database:
+
+- survived = a factor with levels `Yes` and `No`
+- pclass = Passenger Class (1st, 2nd, 3rd). This is a proxy for socio-economic status (SES) 1st ~ Upper; 2nd ~ Middle; 3rd ~ Lower
+
+Suppose we want to test if the proportion of people that survived the sinking of the Titanic differs across passenger classes. To test this hypothesis we select `pclass` as the grouping variable and calculate proportions of `yes` (see `Choose level`) for `survived` (see `Variable (select one)`).
+
+In the `Choose combinations` box select all available entries to conduct pair-wise comparisons across the three passenger class levels. Note that removing all entries will automatically select all combinations. Unless we have an explicit hypothesis for the direction of the effect we should use a two-sided test (i.e., `two.sided`). Our first alternative hypothesis would be 'The proportion of survivors among 1st class passengers was different compared to 2nd class passengers'.
+
+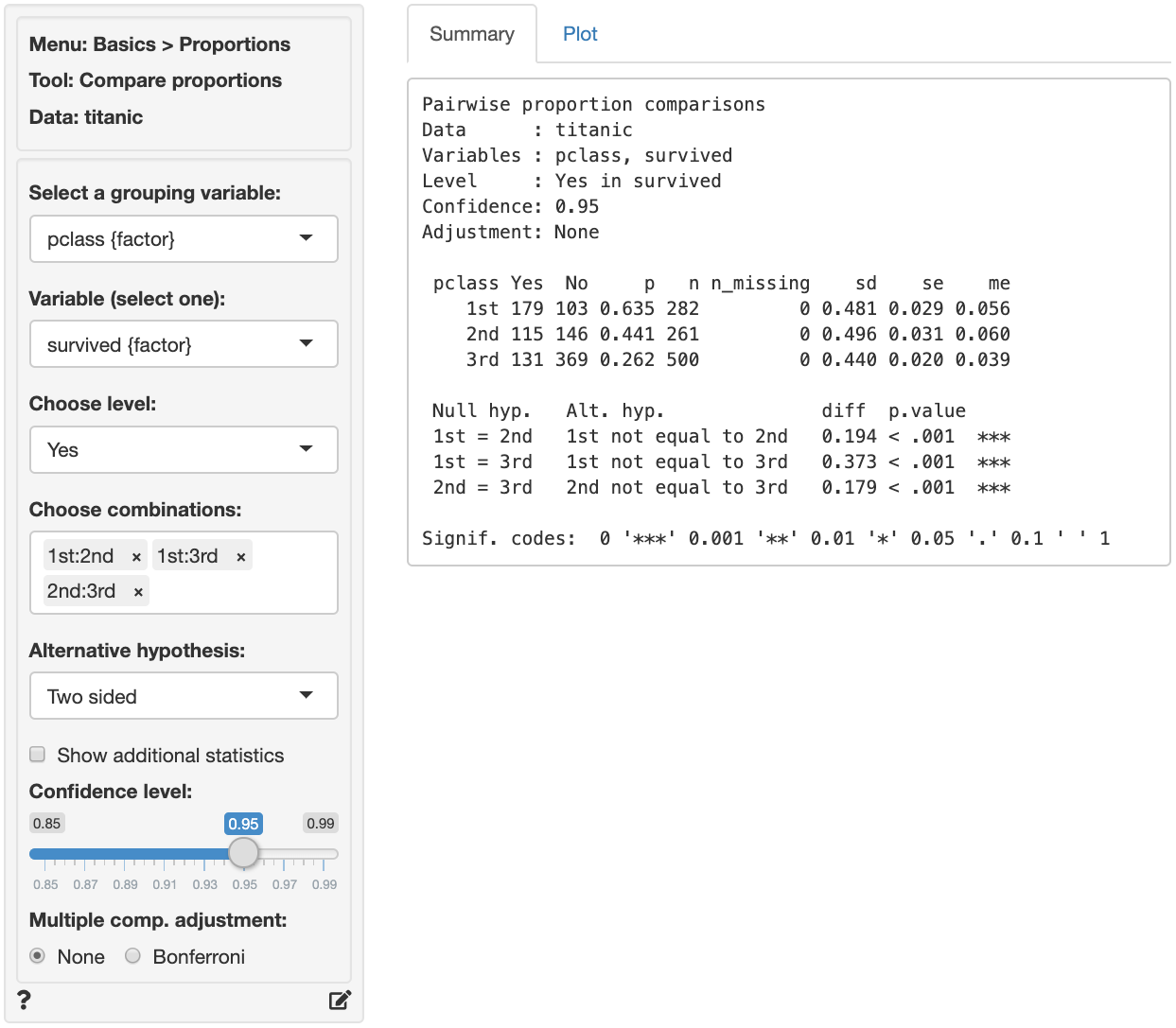
+Pairwise proportion comparisons +Data : titanic +Variables : pclass, survived +Level : Yes in survived +Confidence: 0.95 +Adjustment: None + + pclass Yes No p n n_missing sd se me + 1st 179 103 0.635 282 0 8.086 0.029 0.056 + 2nd 115 146 0.441 261 0 8.021 0.031 0.060 + 3rd 131 369 0.262 500 0 9.832 0.020 0.039 + + Null hyp. Alt. hyp. diff p.value chisq.value df 2.5% 97.5% + 1st = 2nd 1st not equal to 2nd 0.194 < .001 20.576 1 0.112 0.277 *** + 1st = 3rd 1st not equal to 3rd 0.373 < .001 104.704 1 0.305 0.441 *** + 2nd = 3rd 2nd not equal to 3rd 0.179 < .001 25.008 1 0.107 0.250 *** + +Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ++ +* `chisq.value` is the chi-squared statistic associated with `diff` that we can compare to a chi-squared distribution. For additional discussion on how this metric is calculated see the help file in _Basics > Tables > Cross-tabs_. For each combination the equivalent of a 2X2 cross-tab is calculated. +* `df` is the degrees of freedom associated with each statistical test (1). +* `2.5% 97.5%` show the 95% confidence interval around the difference in sample proportions. These numbers provide a range within which the true population difference is likely to fall + +### Testing + +There are three approaches we can use to evaluate the null hypothesis. We will choose a significance level of 0.05.1 Of course, each approach will lead to the same conclusion. + +#### p.value + +Because the p.values are **smaller** than the significance level for each pair-wise comparison we can reject the null hypothesis that the proportions are equal based on the available sample of data. The results suggest that 1st class passengers were more likely to survive the sinking than either 2nd or 3rd class passengers. In turn, the 2nd class passengers were more likely to survive than those in 3rd class. + +#### Confidence interval + +Because zero is **not** contained in any of the confidence intervals we reject the null hypothesis for each evaluated combination of passenger class levels. + +#### Chi-squared values + +Because the calculated chi-squared values (20.576, 104.704, and 25.008) are **larger** than the corresponding _critical_ chi-squared value we reject the null hypothesis for each evaluated combination of passenger class levels. We can obtain the critical chi-squared value by using the probability calculator in the _Basics_ menu. Using the test for 1st versus 2nd class passengers as an example, we find that for a chi-squared distribution with 1 degree of freedom (see `df`) and a confidence level of 0.95 the critical chi-squared value is 3.841. + +
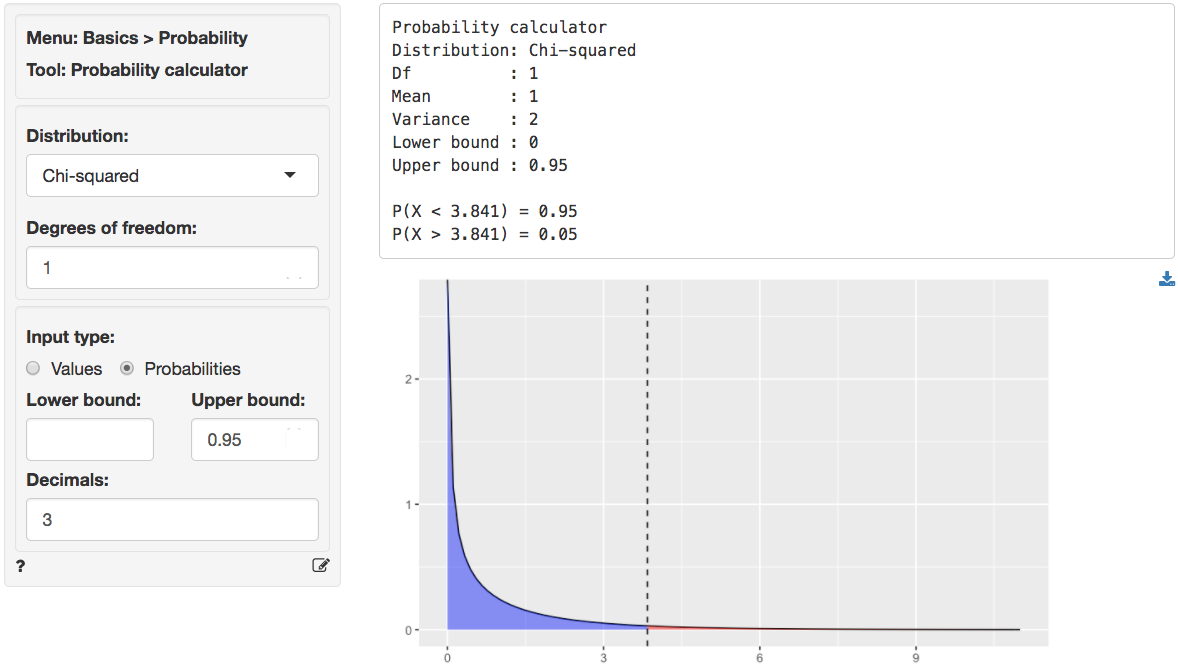
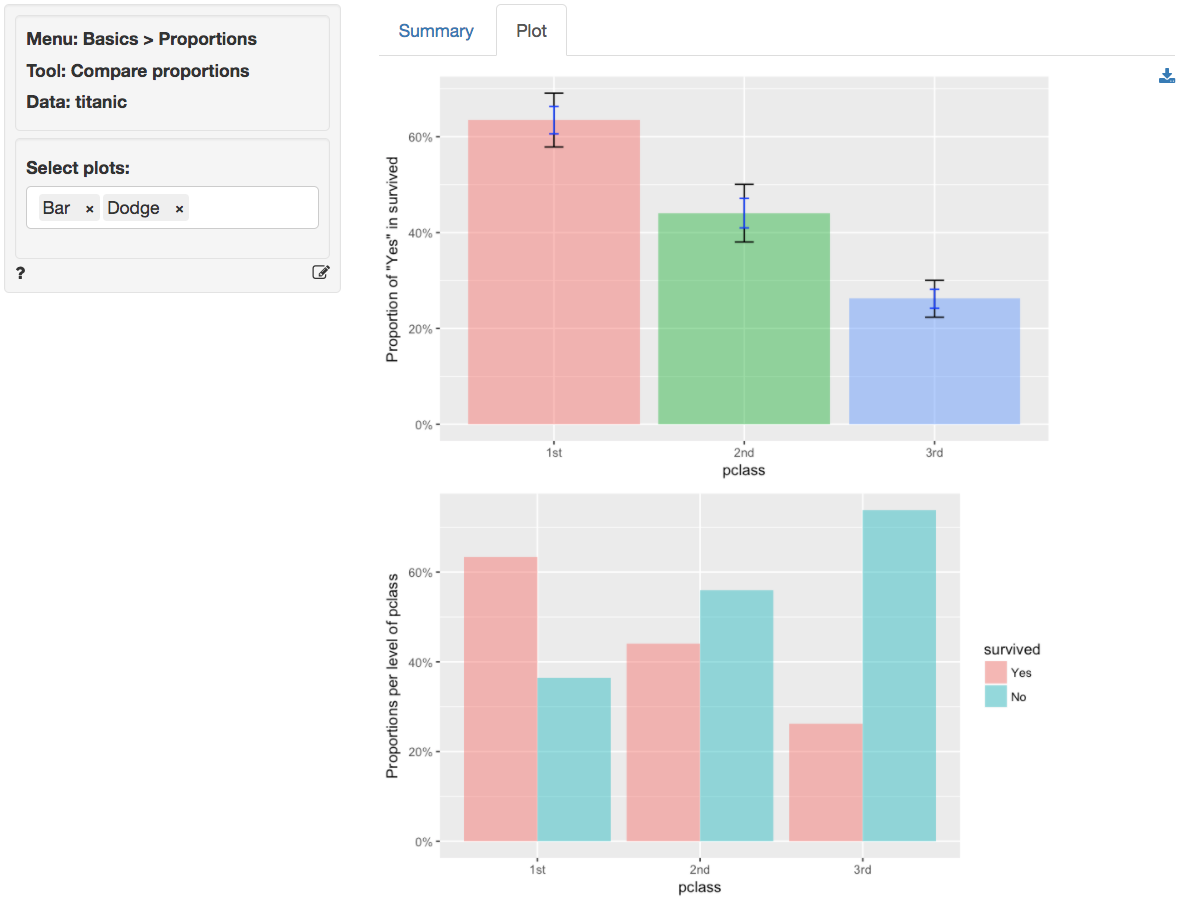
usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+Compare Proportions Hypothesis Test
+
+* This video shows how to conduct a compare proportions hypothesis test
+* Topics List:
+ - Setup a hypothesis test for compare means in Radiant
+ - Use the p.value and confidence interval to evaluate the hypothesis test
+
diff --git a/radiant.basics/vignettes/pkgdown/_correlation.Rmd b/radiant.basics/vignettes/pkgdown/_correlation.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..ac0274bec91c5c28258f831c2c9d8a0996fda127
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/_correlation.Rmd
@@ -0,0 +1,55 @@
+> How correlated are the variables in the data?
+
+Create a correlation matrix of the selected variables. Correlations and p.values are provided for each variable pair. To show only those correlations above a certain (absolute) level, use the correlation cutoff box.
+
+Note: Correlations can be calculated for variables of type `numeric`, `integer`, `date`, and `factor`. When variables of type factor are included the `Adjust for {factor} variables` box should be checked. When correlations are estimated with adjustment, variables that are of type `factor` will be treated as (ordinal) categorical variables and all other variables will be treated as continuous.
+
+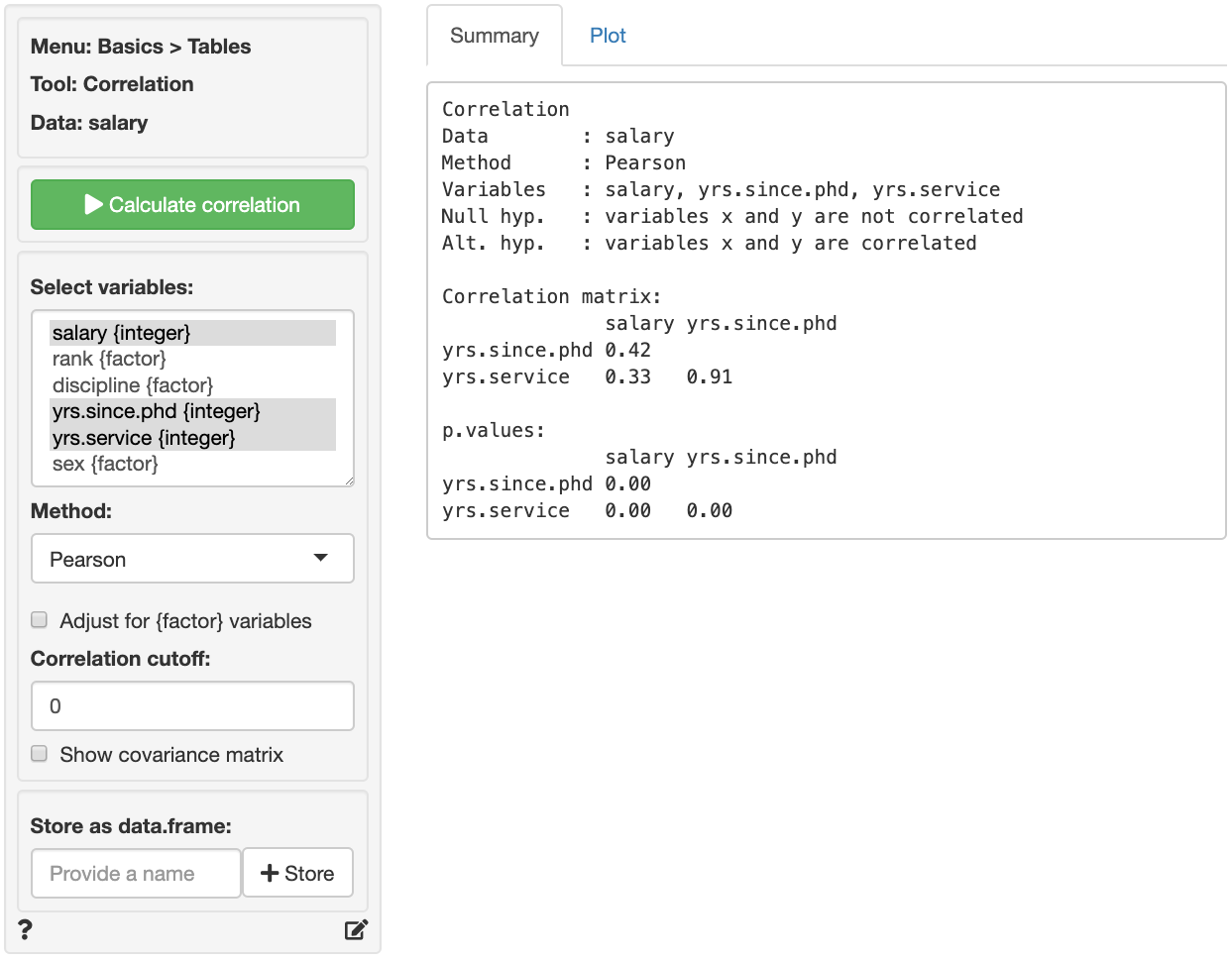
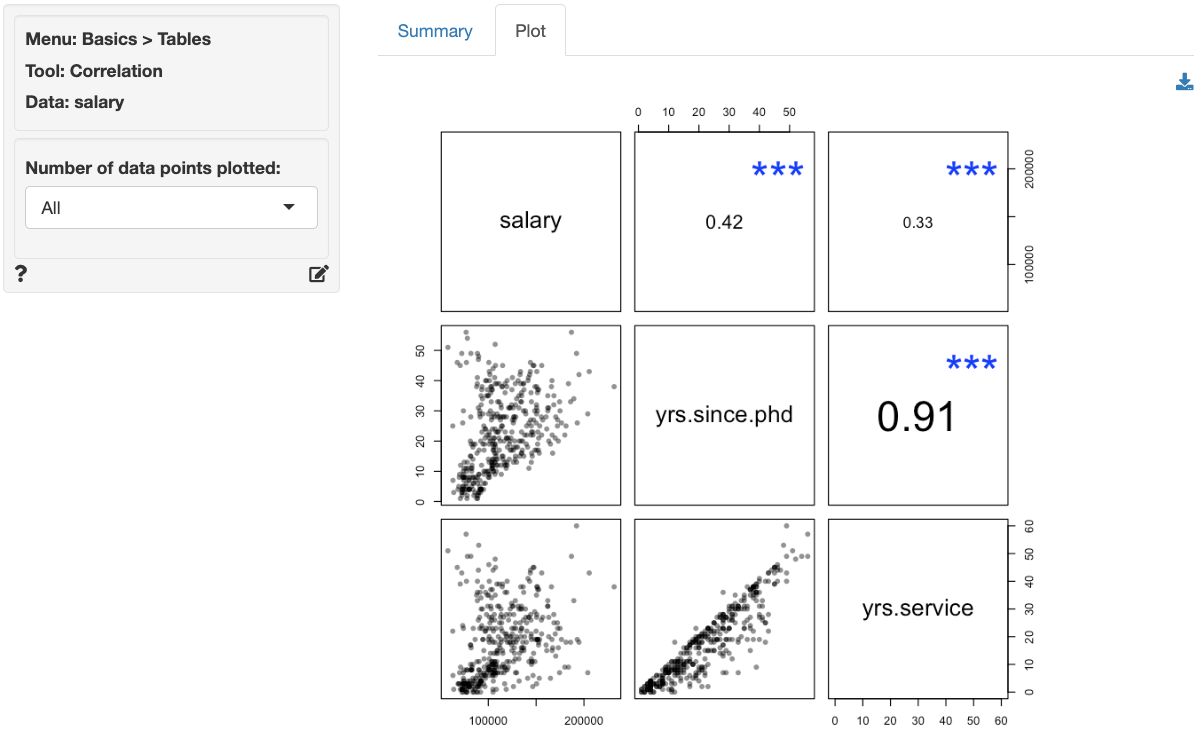
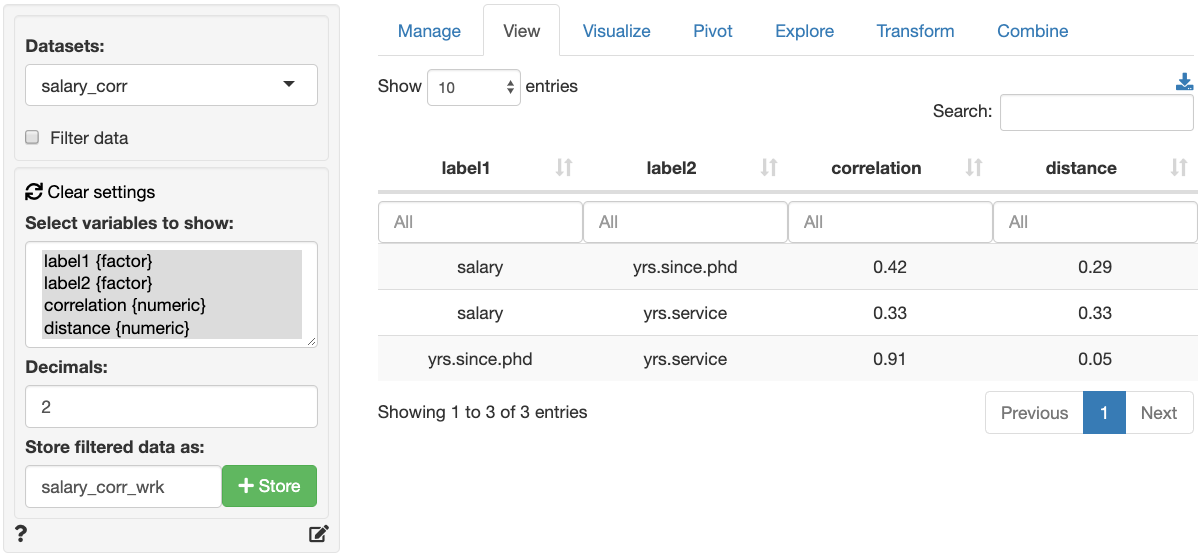
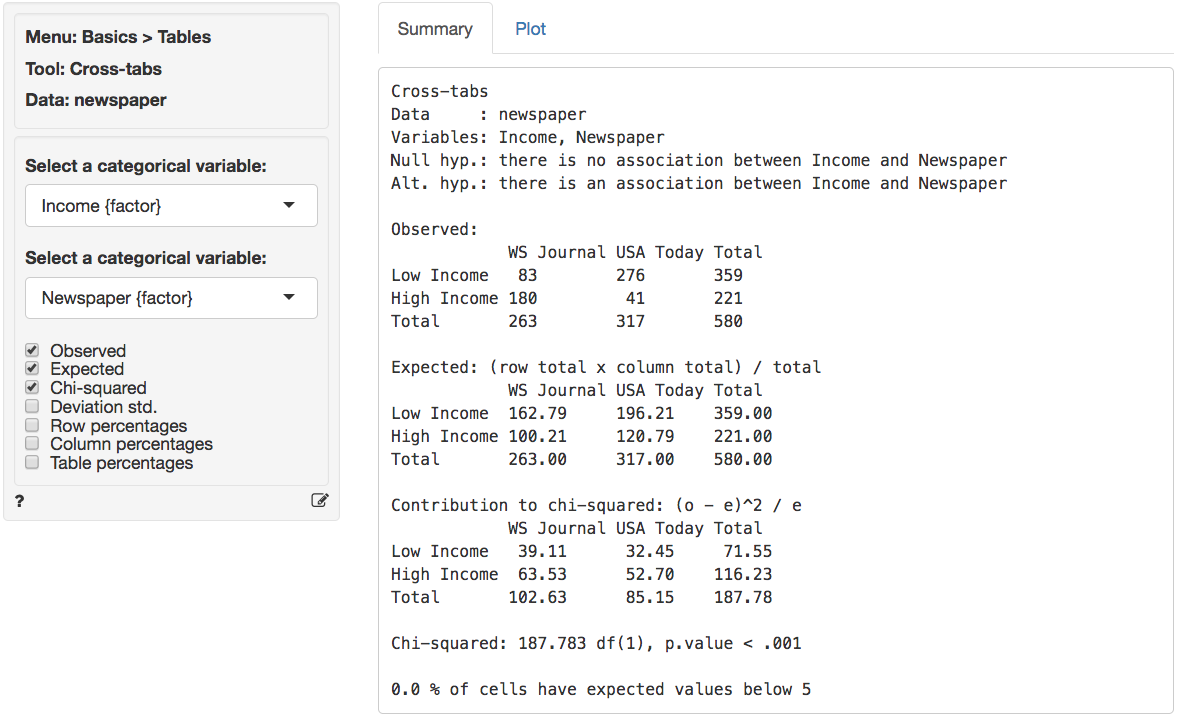

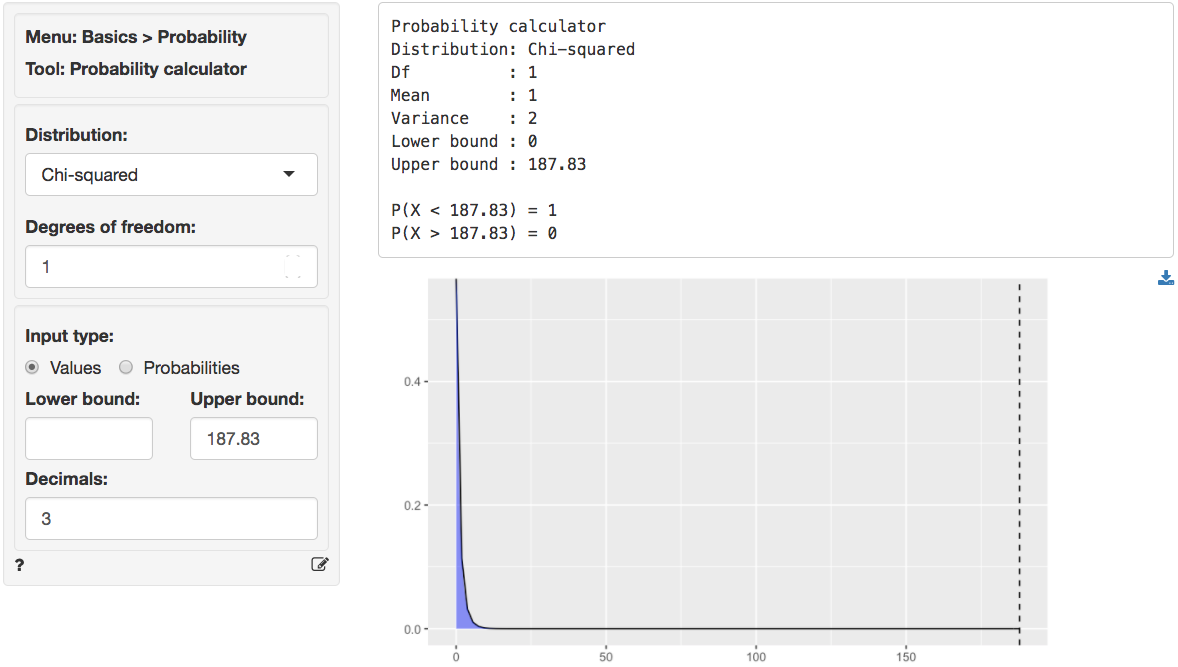
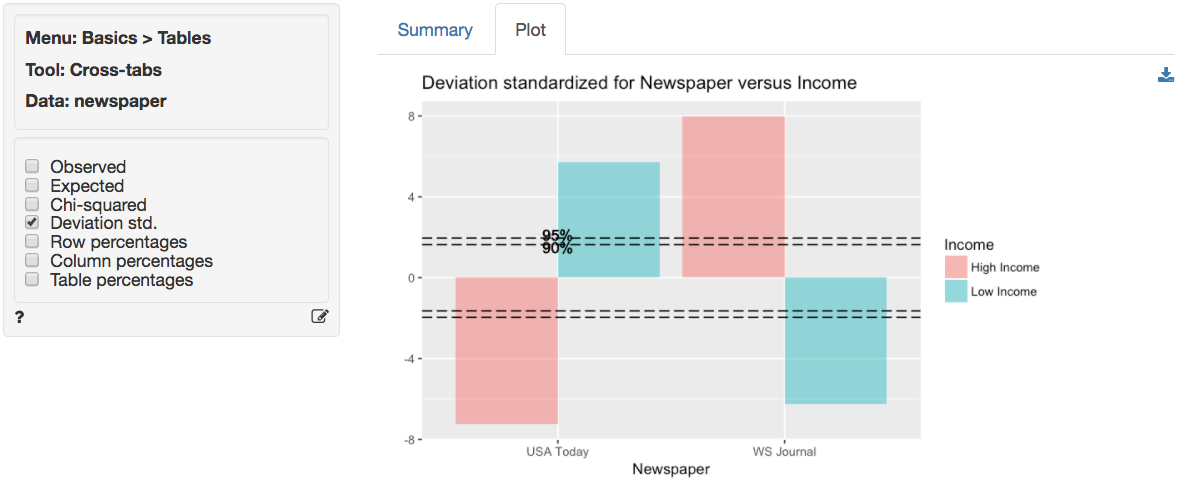
usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+Cross-tabs Hypothesis Test
+
+* This video demonstrates how to investigate associations between two categorical variables by a cross-tabs hypothesis test
+* Topics List:
+ - Setup a hypothesis test for cross-tabs in Radiant
+ - Explain how observed, expected and contribution to chi-squared tables are constructed
+ - Use the p.value and critical value to evaluate the hypothesis test
+
diff --git a/radiant.basics/vignettes/pkgdown/_footer.md b/radiant.basics/vignettes/pkgdown/_footer.md
new file mode 100644
index 0000000000000000000000000000000000000000..05010f02dd76f9e82c3cb8a79ee3cfcec670384d
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/_footer.md
@@ -0,0 +1,2 @@
+
+© Vincent Nijs (2023)  diff --git a/radiant.basics/vignettes/pkgdown/_goodness.Rmd b/radiant.basics/vignettes/pkgdown/_goodness.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..d7339bea9ad2c7c0093db3dff51ca0d2a22501d4
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/_goodness.Rmd
@@ -0,0 +1,46 @@
+> A goodness-of-fit test is used to determine if data from a sample are consistent with a hypothesized distribution
+
+### Example
+
+The data are from a sample of 580 newspaper readers that indicated (1) which newspaper they read most frequently (USA today or Wall Street Journal) and (2) their level of income (Low income vs. High income). The data has three variables: A respondent identifier (id), respondent income (High or Low), and the primary newspaper the respondent reads (USA today or Wall Street Journal).
+
+The data were collected to examine if there is a relationship between income level and choice of newspaper. To ensure the results are generalizable it is important that sample is representative of the population of interest. It is known that in this population the relative share of US today readers is higher than the share of Wall Street Journal readers. The shares should be 55% and 45% respectively. We can use a goodness-of-fit test to examine the following null and alternative hypotheses:
+
+* H0: Readership shares for USA today and Wall Street Journal are 55% and 45% respectively
+* Ha: Readership shares for USA today and Wall Street Journal are not equal to the stated values
+
+If we cannot reject the null hypothesis based on the available sample there is a "good fit" between the observed data and the assumed population shares or probabilities. In Radiant (_Basics > Tables > Goodness of fit_) choose Newspaper as the categorical variable. If we leave the `Probabilities` input field empty (or enter 1/2) we would be testing if the shares are equal. However, to test H0 and Ha we need to enter `0.45 and 0.55` and then press `Enter`. First, compare the observed and expected frequencies. The expected frequencies are calculated assuming H0 is true (i.e., no deviation from the stated shares) as total $\times$ $p$, where $p$ is the share (or probability) assumed for a cell.
+
+
diff --git a/radiant.basics/vignettes/pkgdown/_goodness.Rmd b/radiant.basics/vignettes/pkgdown/_goodness.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..d7339bea9ad2c7c0093db3dff51ca0d2a22501d4
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/_goodness.Rmd
@@ -0,0 +1,46 @@
+> A goodness-of-fit test is used to determine if data from a sample are consistent with a hypothesized distribution
+
+### Example
+
+The data are from a sample of 580 newspaper readers that indicated (1) which newspaper they read most frequently (USA today or Wall Street Journal) and (2) their level of income (Low income vs. High income). The data has three variables: A respondent identifier (id), respondent income (High or Low), and the primary newspaper the respondent reads (USA today or Wall Street Journal).
+
+The data were collected to examine if there is a relationship between income level and choice of newspaper. To ensure the results are generalizable it is important that sample is representative of the population of interest. It is known that in this population the relative share of US today readers is higher than the share of Wall Street Journal readers. The shares should be 55% and 45% respectively. We can use a goodness-of-fit test to examine the following null and alternative hypotheses:
+
+* H0: Readership shares for USA today and Wall Street Journal are 55% and 45% respectively
+* Ha: Readership shares for USA today and Wall Street Journal are not equal to the stated values
+
+If we cannot reject the null hypothesis based on the available sample there is a "good fit" between the observed data and the assumed population shares or probabilities. In Radiant (_Basics > Tables > Goodness of fit_) choose Newspaper as the categorical variable. If we leave the `Probabilities` input field empty (or enter 1/2) we would be testing if the shares are equal. However, to test H0 and Ha we need to enter `0.45 and 0.55` and then press `Enter`. First, compare the observed and expected frequencies. The expected frequencies are calculated assuming H0 is true (i.e., no deviation from the stated shares) as total $\times$ $p$, where $p$ is the share (or probability) assumed for a cell.
+
+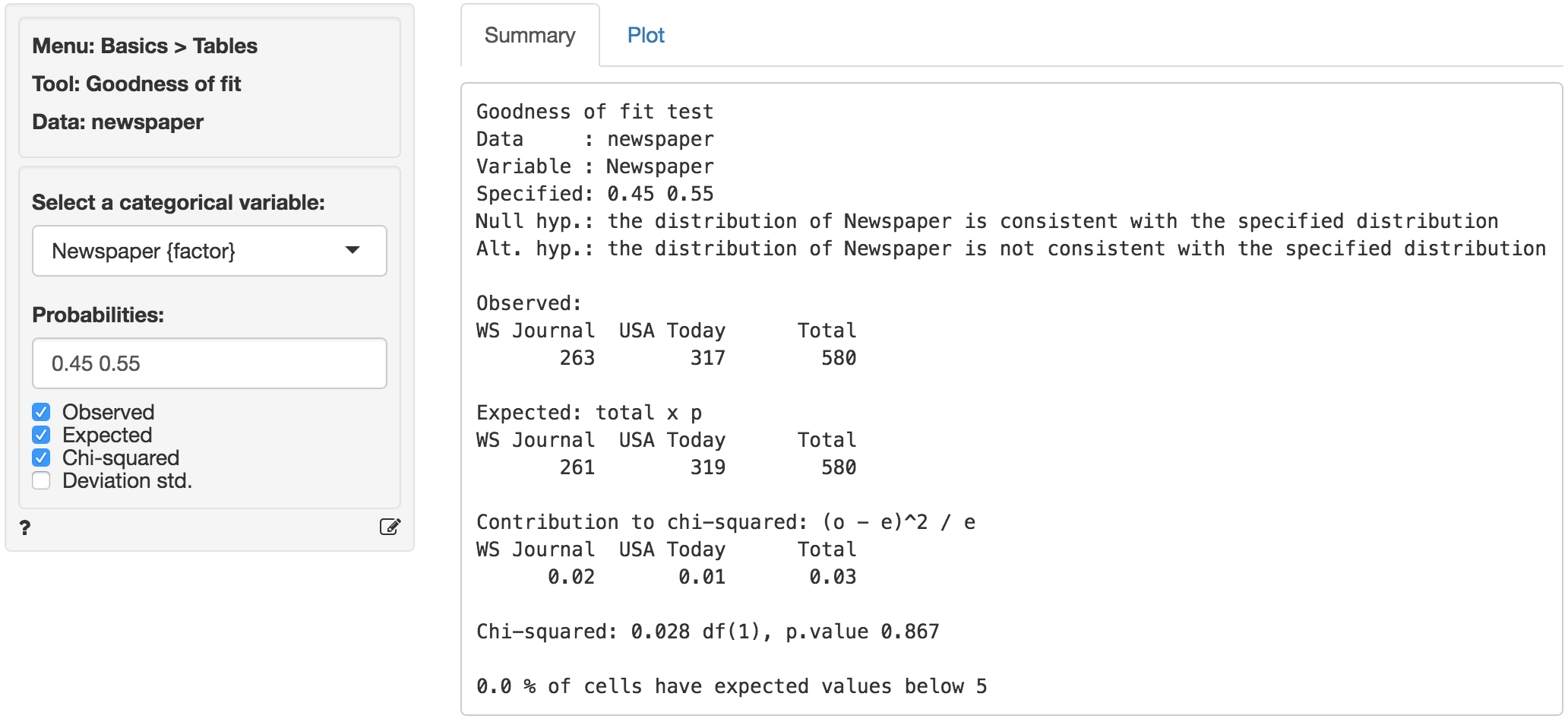

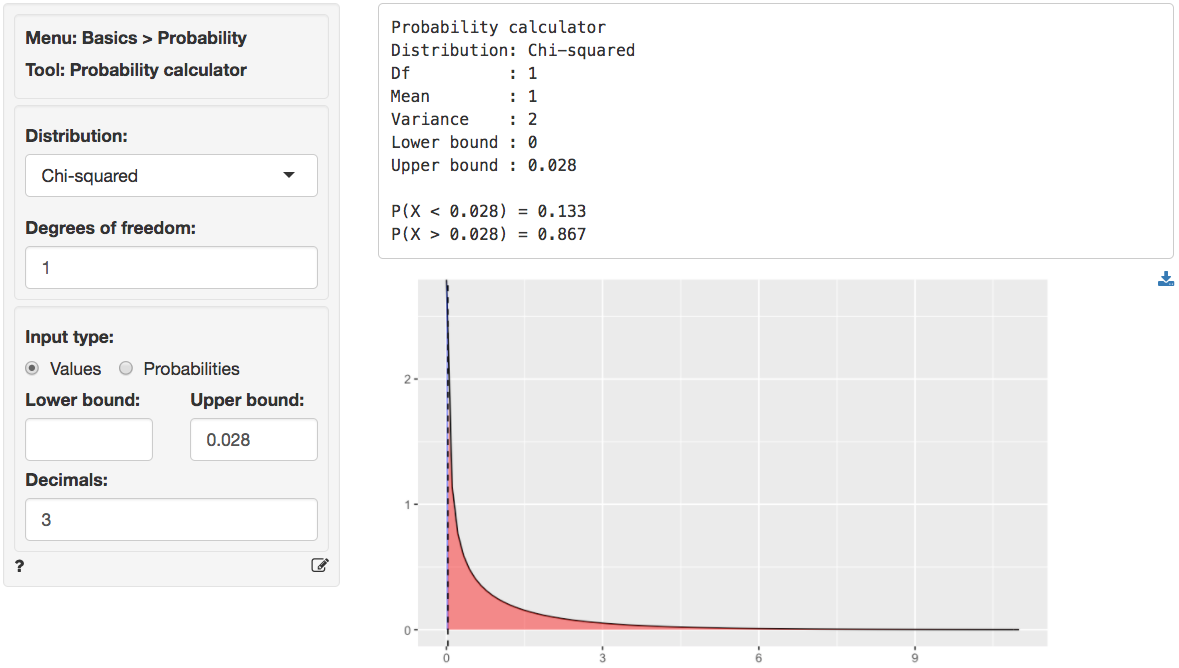
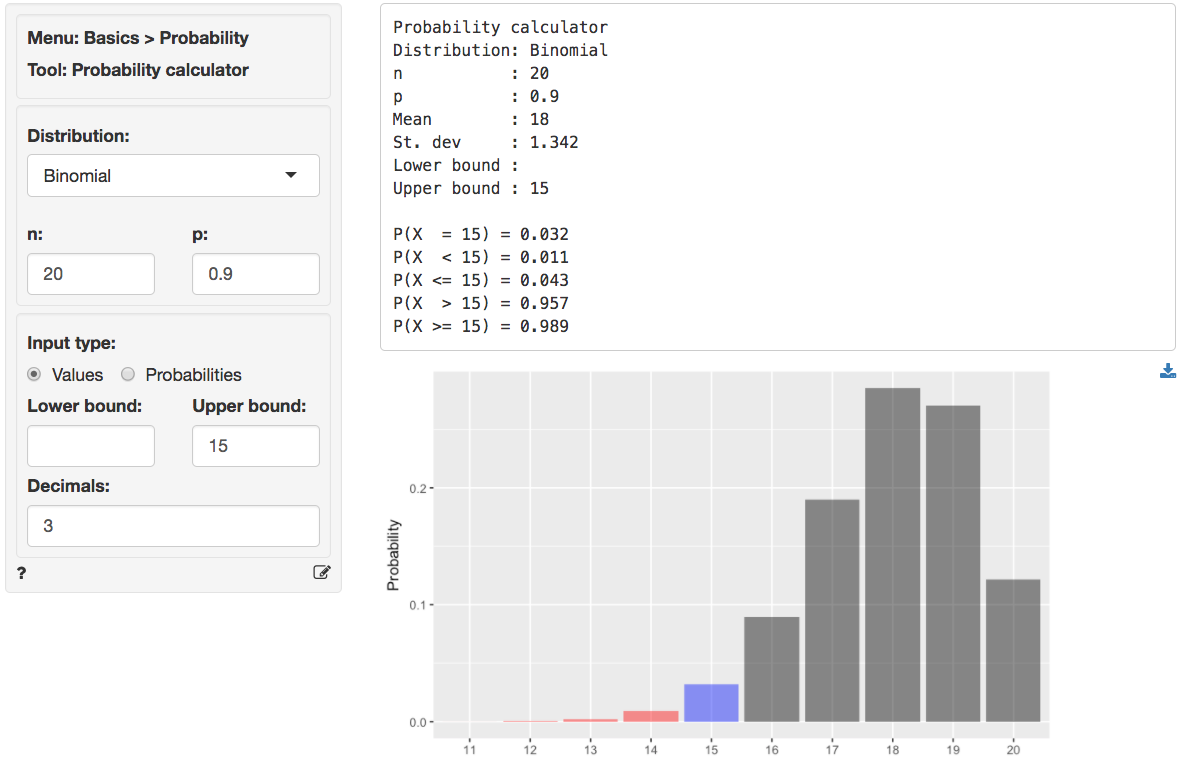
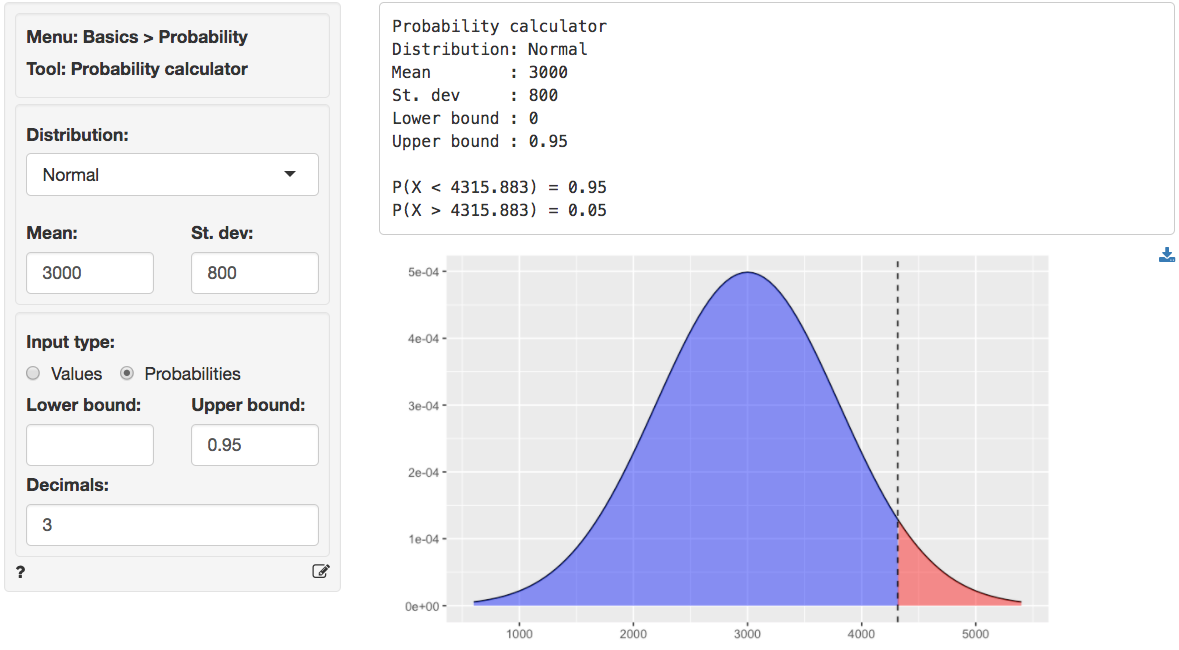
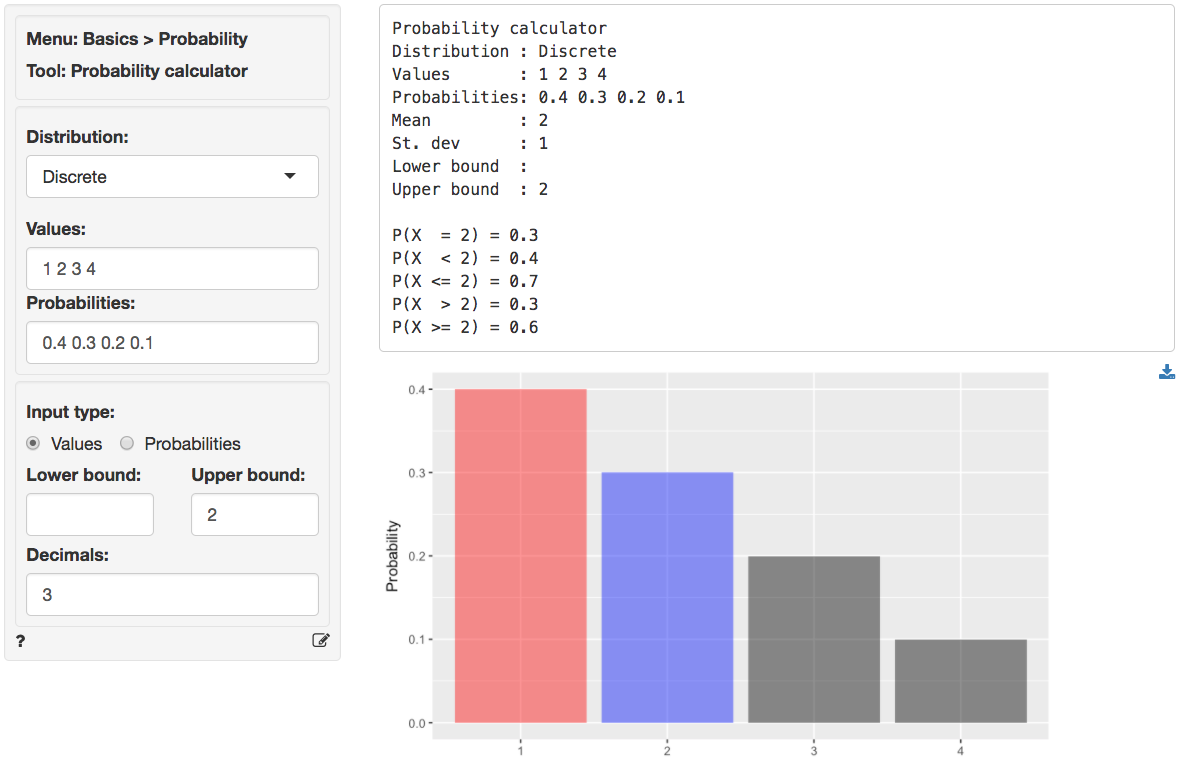
usethis::use_course("https://www.dropbox.com/sh/zw1yuiw8hvs47uc/AABPo1BncYv_i2eZfHQ7dgwCa?dl=1")
+
+Describing the Distribution of a Discrete Random
+ Variable (#1)
+
+* This video shows how to summarize information about a discrete random variable using the probability calculator in Radiant
+* Topics List:
+ - Calculate the mean and variance for a discrete random variable by hand
+ - Calculate the mean, variance, and select probabilities for a discrete random variable in Radiant
+
+Describing Normal and Binomial Distributions in Radiant(#2)
+
+* This video shows how to summarize information about Normal and Binomial distributions using the probability calculator in Radiant
+* Topics List:
+ - Calculate probabilities of a random variable following a Normal distribution in Radiant
+ - Calculate probabilities of a random variable following a Binomial distribution by hand
+ - Calculate probabilities of a random variable following a Binomial distribution in Radiant
+
+Describing Uniform and Binomial Distributions in Radiant(#3)
+
+* This video shows how to summarize information about Uniform and Binomial distributions using the probability calculator in Radiant
+* Topics List:
+ - Calculate probabilities of a random variable following a Uniform distribution in Radiant
+ - Calculate probabilities of a random variable following a Binomial distribution in Radiant
+
+Providing Probability Bounds(#4)
+
+* This video demonstrates how to provide probability bounds in Radiant
+* Topics List:
+ - Use probabilities as input type
+ - Round up the cutoff value
diff --git a/radiant.basics/vignettes/pkgdown/_single_mean.Rmd b/radiant.basics/vignettes/pkgdown/_single_mean.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..80228a653a68b98eb05e8aadde052ea8b3e382aa
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/_single_mean.Rmd
@@ -0,0 +1,82 @@
+> Compare a single mean to the mean value in the population
+
+The single mean (or one-sample) t-test is used to compare the mean of a variable in a sample of data to a (hypothesized) mean in the population from which our sample data are drawn. This is important because we seldom have access to data for an entire population. The hypothesized value in the population is specified in the `Comparison value` box.
+
+We can perform either a one-sided test (i.e., `less than` or `greater than`) or a two-sided test (see the `Alternative hypothesis` dropdown). We use one-sided tests to evaluate if the available data provide evidence that the sample mean is larger (or smaller) than the comparison value (i.e., the population value in the null-hypothesis).
+
+## Example
+
+We have access to data from a random sample of grocery stores in the UK. Management will consider entering this market if consumer demand for the product category exceeds 100M units, or, approximately, 1750 units per store. The average demand per store in the sample is equal to 1953. While this number is larger than 1750 we need to determine if the difference could be attributed to sampling error.
+
+You can find the information on unit sales in each of the sample stores in the **demand\_uk.rda** data set. The data set contains two variables, `store_id` and `demand_uk`. Our null-hypothesis is that the average store demand in the UK is equal to 1750 unit so we enter that number into the `Comparison value` box. We choose the `Greater than` option from the `Alternative hypothesis` drop-down because we want to determine if the available data provides sufficient evidence to reject the null-hypothesis favor of the alternative that average store demand in the UK is **larger than 1750**.
+
+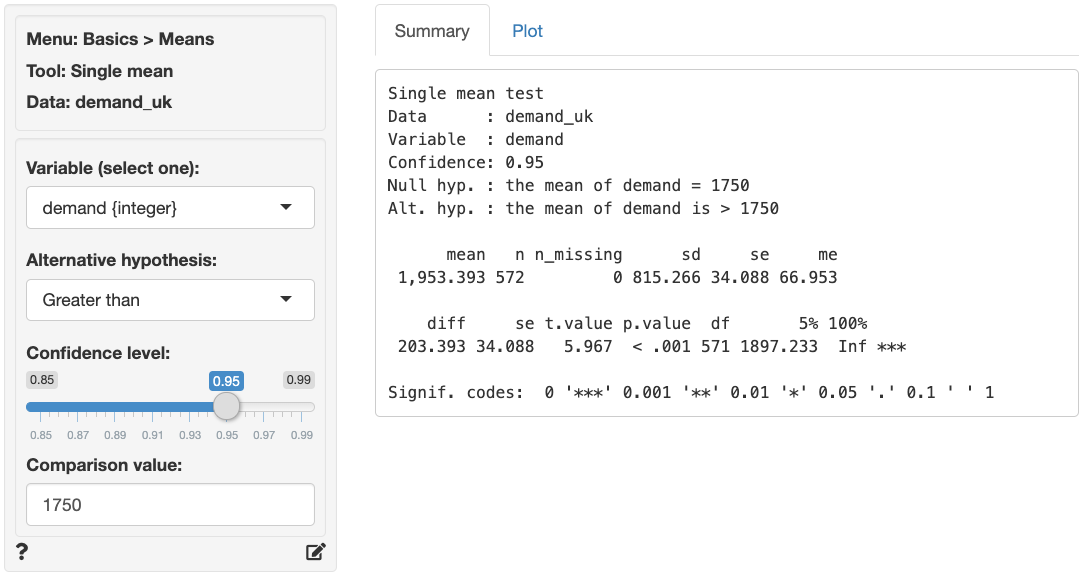
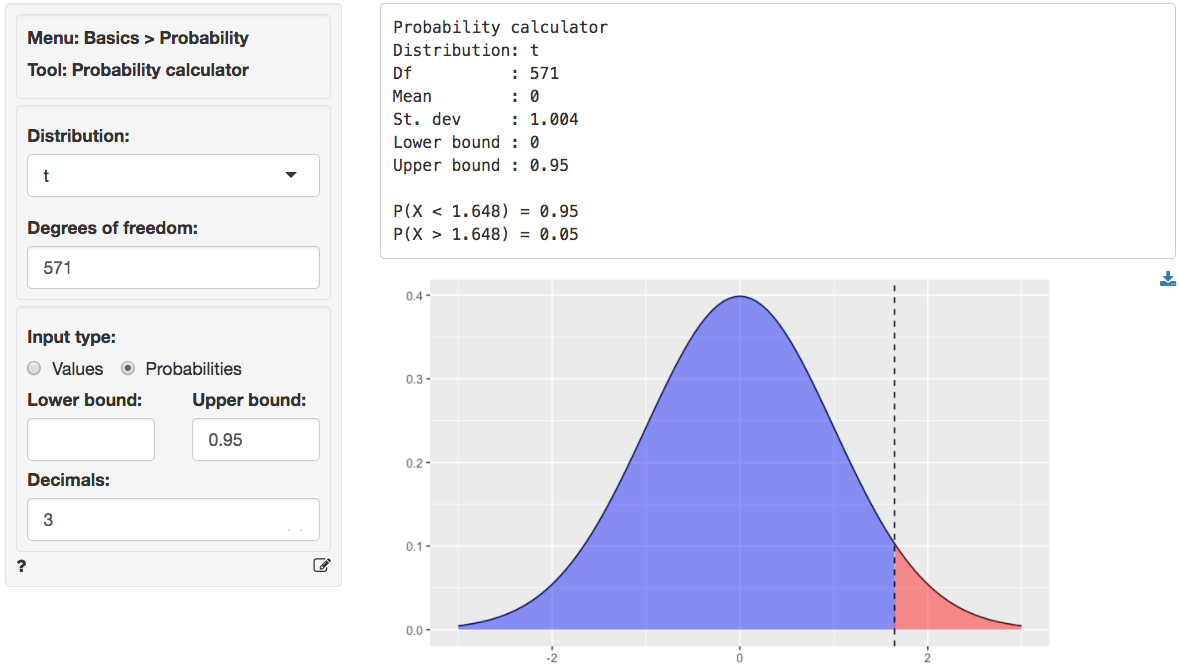
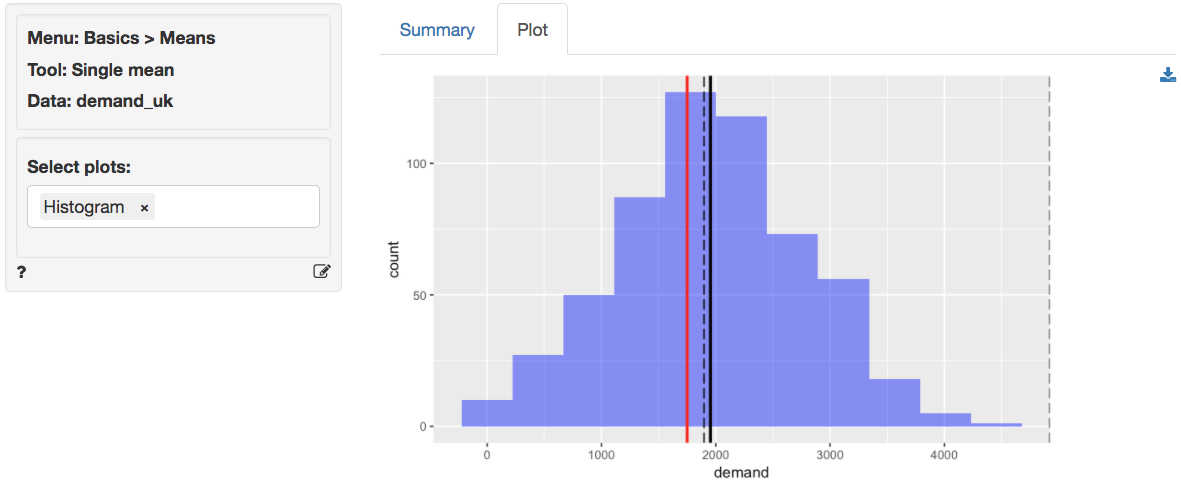
usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+Single Mean Hypothesis Test
+
+* This video shows how to test a hypothesis about a single sample mean versus a population mean
+* Topics List:
+ - Calculate summary statistics for a sample
+ - Setup a hypothesis test for a single mean in Radiant
+ - Use the p.value, confidence interval, or critical value to evaluate the hypothesis test
diff --git a/radiant.basics/vignettes/pkgdown/_single_prop.Rmd b/radiant.basics/vignettes/pkgdown/_single_prop.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..f113a8577dfc5aed15eb8614b87726cdc1e7e385
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/_single_prop.Rmd
@@ -0,0 +1,87 @@
+> Compare a single proportion to the population proportion
+
+The single proportion (or one-sample) binomial test is used to compare a proportion of responses or values in a sample of data to a (hypothesized) proportion in the population from which our sample data are drawn. This is important because we seldom have access to data for an entire population. The hypothesized value in the population is specified in the `Comparison value` box.
+
+We can perform either a one-sided test (i.e., `less than` or `greater than`) or a two-sided test (see the `Alternative hypothesis` dropdown). We use one-sided tests to evaluate if the available data provide evidence that a sample proportion is larger (or smaller) than the comparison value (i.e., the population value in the null-hypothesis).
+
+## Example
+
+A car manufacturer conducted a study by randomly sampling and interviewing 1,000 consumers in a new target market. The goal of the study was to determine if consumers would consider purchasing this brand of car.
+
+Management has already determined that the company will enter this segment. However, if brand preference is lower than 10% additional resources will be committed to advertising and sponsorship in an effort to enhance brand awareness among the target consumers. In the sample, 93 consumers exhibited what the company considered strong brand liking.
+
+You can find information on the responses by survey participants in the **consider.rda** data set. The data set contains two variables, `id` and `consider`.
+
+Our null-hypothesis is that the proportion of consumers that would consider the car brand for a future purchase is equal to 10%. Select the `consider` variable from the `Variable` dropdown. To evaluate the proportion of `yes` responses in the sample select `yes` from the `Choose level` dropdown.
+
+Choose the `Less than` option from the `Alternative hypothesis` drop-down to determine if the available data provides sufficient evidence to reject the null-hypothesis in favor of the alternative that the proportion of consumers that will consider the brand is **less than 10%**.
+
+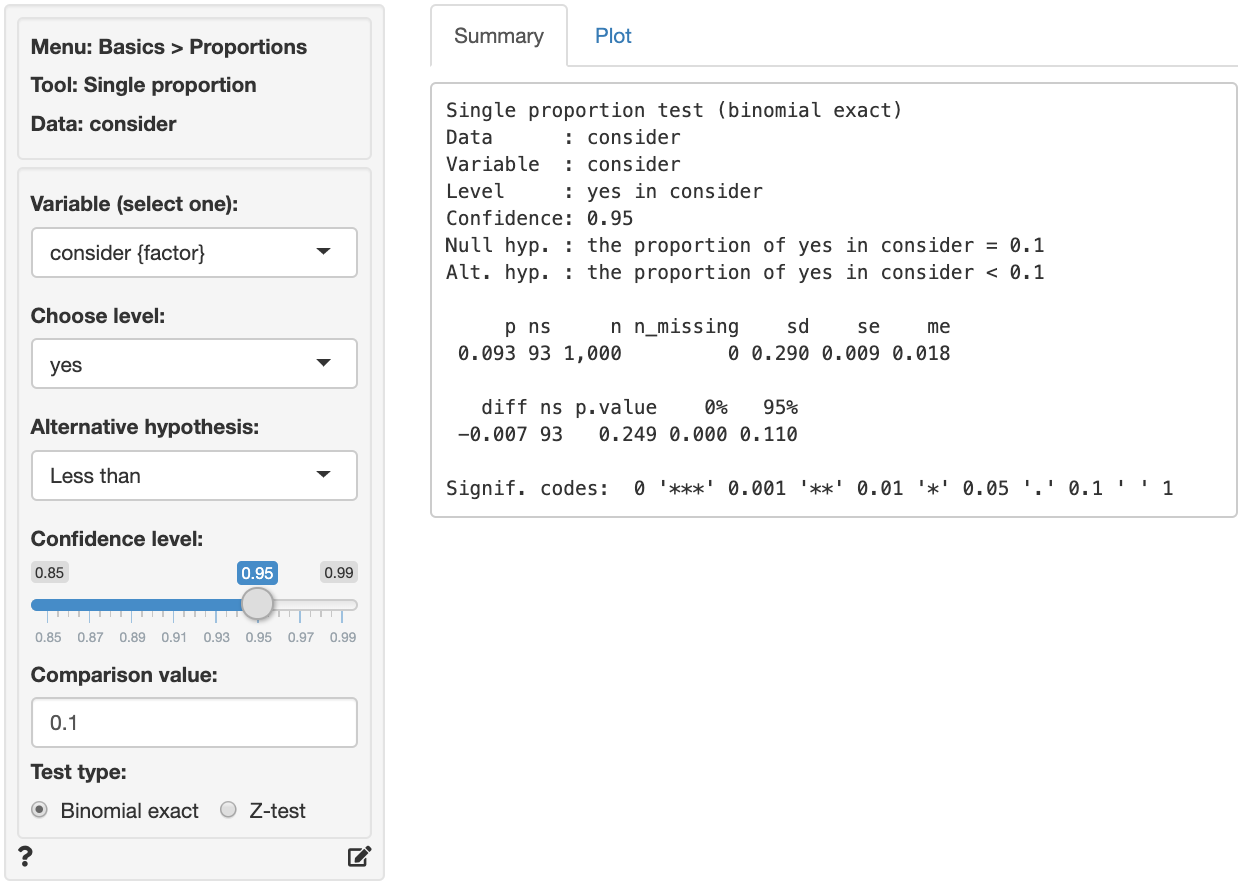
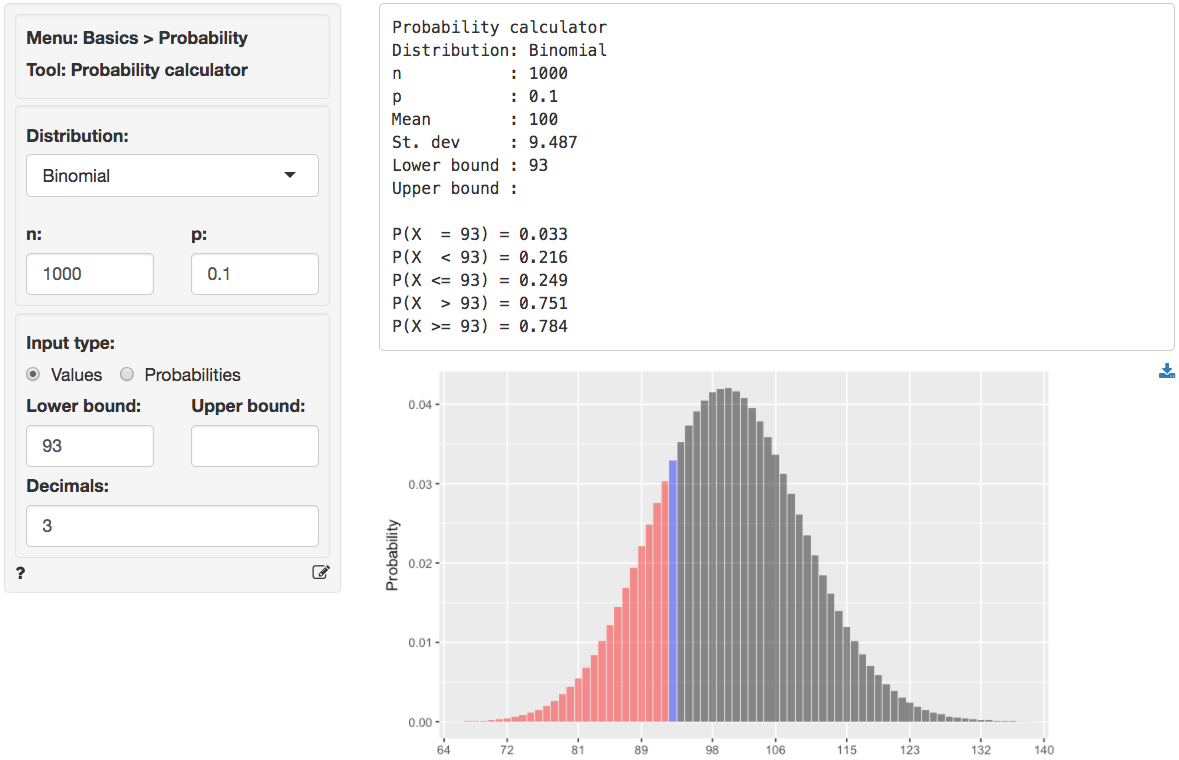
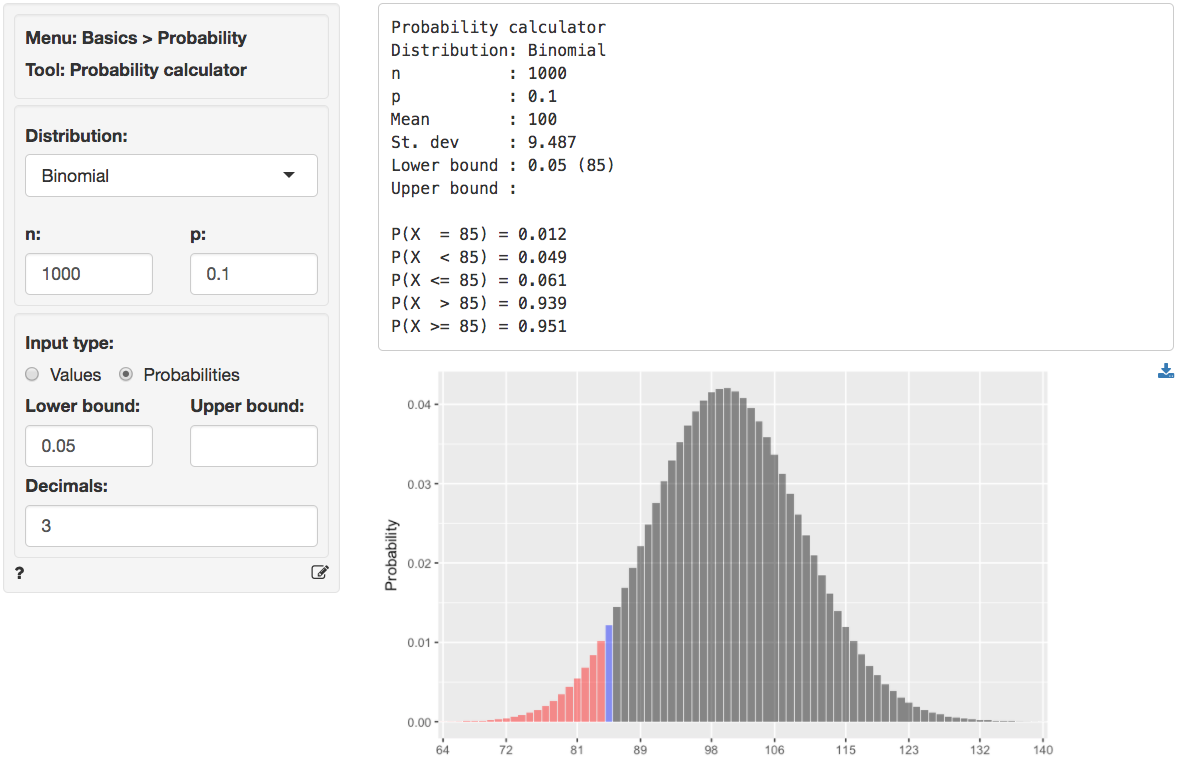
usethis::use_course("https://www.dropbox.com/sh/0xvhyolgcvox685/AADSppNSIocrJS-BqZXhD1Kna?dl=1")
+
+Single Proportion Hypothesis Test
+
+* This video shows how to test a hypothesis about a single sample proportion versus a population proportion
+* Topics List:
+ - Setup a hypothesis test for a single proportion in Radiant
+ - Use the p.value, confidence interval, or critical value to evaluate the hypothesis test
diff --git a/radiant.basics/vignettes/pkgdown/clt.Rmd b/radiant.basics/vignettes/pkgdown/clt.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..38408943fc720e594492528479bf9b5478bfac4b
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/clt.Rmd
@@ -0,0 +1,10 @@
+---
+title: "Basics > Central Limit Theorem"
+author: "Vincent R. Nijs, Rady School of Management (UCSD)"
+---
+
+```{r child = "_clt.Rmd"}
+```
+
+```{r child = "_footer.md"}
+```
diff --git a/radiant.basics/vignettes/pkgdown/compare_means.Rmd b/radiant.basics/vignettes/pkgdown/compare_means.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..04662318a4b3d8eb335cee6bd65061a2d7787342
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/compare_means.Rmd
@@ -0,0 +1,11 @@
+---
+title: "Basics > Compare means"
+author: "Vincent R. Nijs, Rady School of Management (UCSD)"
+---
+
+```{r child = "_compare_means.Rmd"}
+```
+
+```{r child = "_footer.md"}
+```
+
diff --git a/radiant.basics/vignettes/pkgdown/compare_props.Rmd b/radiant.basics/vignettes/pkgdown/compare_props.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..6f4f74fd8ecaf6e40cc79259ac0dfdbe6340a188
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/compare_props.Rmd
@@ -0,0 +1,10 @@
+---
+title: "Basics > Compare proportions"
+author: "Vincent R. Nijs, Rady School of Management (UCSD)"
+---
+
+```{r child = "_compare_props.Rmd"}
+```
+
+```{r child = "_footer.md"}
+```
diff --git a/radiant.basics/vignettes/pkgdown/correlation.Rmd b/radiant.basics/vignettes/pkgdown/correlation.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..5824c19788b0723e93e362ce4ea51790357905b6
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/correlation.Rmd
@@ -0,0 +1,11 @@
+---
+title: "Basics > Correlation"
+author: "Vincent R. Nijs, Rady School of Management (UCSD)"
+---
+
+```{r child = "_correlation.Rmd"}
+```
+
+```{r child = "_footer.md"}
+```
+
diff --git a/radiant.basics/vignettes/pkgdown/cross_tabs.Rmd b/radiant.basics/vignettes/pkgdown/cross_tabs.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..2080fe8fdfaeb145539fb98c2489c65138ee0a45
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/cross_tabs.Rmd
@@ -0,0 +1,11 @@
+---
+title: "Basics > Cross-tabs"
+author: "Vincent R. Nijs, Rady School of Management (UCSD)"
+---
+
+```{r child = "_cross_tabs.Rmd"}
+```
+
+```{r child = "_footer.md"}
+```
+
diff --git a/radiant.basics/vignettes/pkgdown/goodness.Rmd b/radiant.basics/vignettes/pkgdown/goodness.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..ed2c1f2465339ed4265783cf57bbe2303dc05c9c
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/goodness.Rmd
@@ -0,0 +1,11 @@
+---
+title: "Basics > Goodness of fit"
+author: "Vincent R. Nijs, Rady School of Management (UCSD)"
+---
+
+```{r child = "_goodness.Rmd"}
+```
+
+```{r child = "_footer.md"}
+```
+
diff --git a/radiant.basics/vignettes/pkgdown/images/by-nc-sa.png b/radiant.basics/vignettes/pkgdown/images/by-nc-sa.png
new file mode 100644
index 0000000000000000000000000000000000000000..76eb5da461b41405c500a557253eec5f65169519
Binary files /dev/null and b/radiant.basics/vignettes/pkgdown/images/by-nc-sa.png differ
diff --git a/radiant.basics/vignettes/pkgdown/prob_calc.Rmd b/radiant.basics/vignettes/pkgdown/prob_calc.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..ba1165ed7089803c29bba19a05d44b3729aaa203
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/prob_calc.Rmd
@@ -0,0 +1,10 @@
+---
+title: "Basics > Probability calculator"
+author: "Vincent R. Nijs, Rady School of Management (UCSD)"
+---
+
+```{r child = "_prob_calc.Rmd"}
+```
+
+```{r child = "_footer.md"}
+```
diff --git a/radiant.basics/vignettes/pkgdown/single_mean.Rmd b/radiant.basics/vignettes/pkgdown/single_mean.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..630db385a8eeeb4d284798784ab988cc97f0e812
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/single_mean.Rmd
@@ -0,0 +1,11 @@
+---
+title: "Basics > Single mean"
+author: "Vincent R. Nijs, Rady School of Management (UCSD)"
+---
+
+```{r child = "_single_mean.Rmd"}
+```
+
+```{r child = "_footer.md"}
+```
+
diff --git a/radiant.basics/vignettes/pkgdown/single_prop.Rmd b/radiant.basics/vignettes/pkgdown/single_prop.Rmd
new file mode 100644
index 0000000000000000000000000000000000000000..57db4a3c26fb09a312f92a5b32e9bbfc745b3af0
--- /dev/null
+++ b/radiant.basics/vignettes/pkgdown/single_prop.Rmd
@@ -0,0 +1,11 @@
+---
+title: "Basics > Single proportion"
+author: "Vincent R. Nijs, Rady School of Management (UCSD)"
+---
+
+```{r child = "_single_prop.Rmd"}
+```
+
+```{r child = "_footer.md"}
+```
+
diff --git a/radiant.data b/radiant.data
deleted file mode 160000
index 8981e238837b6bcc95e2811c9509162ee54b3088..0000000000000000000000000000000000000000
--- a/radiant.data
+++ /dev/null
@@ -1 +0,0 @@
-Subproject commit 8981e238837b6bcc95e2811c9509162ee54b3088
diff --git a/radiant.data/.Rbuildignore b/radiant.data/.Rbuildignore
new file mode 100644
index 0000000000000000000000000000000000000000..8efef6658f700d1c7b2363b8bd8aee42146e4d29
--- /dev/null
+++ b/radiant.data/.Rbuildignore
@@ -0,0 +1,18 @@
+^CRAN-RELEASE$
+^.*\.Rproj$
+^\.Rproj\.user$
+^inst/rstudio$
+^build$
+^docs$
+^vignettes$
+^\.travis\.yml$
+_pkgdown.yml
+cran-comments.md
+coverage.R
+^tests/testthat/data$
+.vscode
+radiant.data.code-workspace
+^CRAN-SUBMISSION$
+R/app.R
+^.codespellrc$
+^.github$
diff --git a/radiant.data/.codespellrc b/radiant.data/.codespellrc
new file mode 100644
index 0000000000000000000000000000000000000000..baeecfca45067c8c5bda6b3b4d832f8e5d698926
--- /dev/null
+++ b/radiant.data/.codespellrc
@@ -0,0 +1,5 @@
+[codespell]
+skip = .git,*.pdf,*.svg,*.min.js,*.csv,*.html
+# rady - server/domain name
+# nd, isTs - variable names
+ignore-words-list = rady,nd,ists
diff --git a/radiant.data/.github/workflows/codespell.yml b/radiant.data/.github/workflows/codespell.yml
new file mode 100644
index 0000000000000000000000000000000000000000..5768d7c63672e68e60791ca6828d50d76be35e61
--- /dev/null
+++ b/radiant.data/.github/workflows/codespell.yml
@@ -0,0 +1,19 @@
+---
+name: Codespell
+
+on:
+ push:
+ branches: [master]
+ pull_request:
+ branches: [master]
+
+jobs:
+ codespell:
+ name: Check for spelling errors
+ runs-on: ubuntu-latest
+
+ steps:
+ - name: Checkout
+ uses: actions/checkout@v3
+ - name: Codespell
+ uses: codespell-project/actions-codespell@v1
diff --git a/radiant.data/.gitignore b/radiant.data/.gitignore
new file mode 100644
index 0000000000000000000000000000000000000000..20d099f783e75a214cff301b29258ae11559db30
--- /dev/null
+++ b/radiant.data/.gitignore
@@ -0,0 +1,12 @@
+.Rproj.user
+.Rhistory
+.Rapp.history
+.RData
+.Ruserdata
+radiant.data.Rproj
+.DS_Store
+revdep/
+.vscode
+radiant-data.dcf
+radiant-data.Rproj
+R/app.R
diff --git a/radiant.data/.travis.yml b/radiant.data/.travis.yml
new file mode 100644
index 0000000000000000000000000000000000000000..ac61638545836ea37ea3f6a16636496886cf5290
--- /dev/null
+++ b/radiant.data/.travis.yml
@@ -0,0 +1,25 @@
+language: r
+cache: packages
+r:
+ - oldrel
+ - release
+ - devel
+warnings_are_errors: true
+sudo: required
+dist: bionic
+
+r_packages:
+ - devtools
+
+after_success:
+ - Rscript -e 'pkgdown::build_site()'
+
+## based on https://www.datacamp.com/community/tutorials/cd-package-docs-pkgdown-travis
+deploy:
+ provider: pages
+ skip-cleanup: true
+ github-token: $GITHUB_PAT
+ keep-history: true
+ local-dir: docs
+ on:
+ branch: master
diff --git a/radiant.data/COPYING b/radiant.data/COPYING
new file mode 100644
index 0000000000000000000000000000000000000000..0fb9090d488e333fb8eb8461a561f215990278b3
--- /dev/null
+++ b/radiant.data/COPYING
@@ -0,0 +1,727 @@
+The radiant.data package is licensed to you under the AGPLv3, the terms of
+which are included below. The help files for radiant.data are licensed under the creative commons attribution and share-alike license [CC-BY-SA].
+
+Radiant code license
+--------------------------------------------------------------------------------------------
+ GNU AFFERO GENERAL PUBLIC LICENSE
+ Version 3, 19 November 2007
+
+ Copyright (C) 2007 Free Software Foundation, Inc. \nBy: ", paste0(by, collapse = ", ")) + } else { + x <- get(type, envir = as.environment("package:dplyr"))(x, y) + madd <- "" + } + + ## return error message as needed + if (is.character(x)) { + return(x) + } + + mess <- paste0( + "## Combined\n\nDatasets: ", x_name, " and ", y_name, + " (", type, ")", madd, "\nOn: ", lubridate::now(), "\n\n", x_descr, + ifelse(!is.empty(data_filter), paste0("\n\n**Data filter:** ", data_filter), ""), + ifelse(!is.empty(arr), paste0("\n\n**Data arrange:** ", make_arrange_cmd(arr)), ""), + ifelse(!is.empty(rows), paste0("\n\n**Data slice:** ", rows), ""), + "\n\n", y_descr + ) + + set_attr(x, "description", mess) +} \ No newline at end of file diff --git a/radiant.data/R/deprecated.R b/radiant.data/R/deprecated.R new file mode 100644 index 0000000000000000000000000000000000000000..73bc25108cd830edbd355c72c8859efaa4d33749 --- /dev/null +++ b/radiant.data/R/deprecated.R @@ -0,0 +1,118 @@ +#' Deprecated function(s) in the radiant.data package +#' +#' These functions are provided for compatibility with previous versions of +#' radiant but will be removed +#' @rdname radiant.data-deprecated +#' @name radiant.data-deprecated +#' @param ... Parameters to be passed to the updated functions +#' @export mean_rm median_rm min_rm max_rm sd_rm var_rm sum_rm getdata filterdata combinedata viewdata toFct fixMS getsummary Search formatnr formatdf rounddf getclass is_numeric +#' @aliases mean_rm median_rm min_rm max_rm sd_rm var_rm sum_rm getdata filterdata combinedata viewdata toFct fixMS getsummary Search formatnr formatdf rounddf getclass is_numeric +#' @section Details: +#' \itemize{ +#' \item Replace \code{mean_rm} by \code{\link{mean}} +#' \item Replace \code{median_rm} by \code{\link{median}} +#' \item Replace \code{min_rm} by \code{\link{min}} +#' \item Replace \code{max_rm} by \code{\link{max}} +#' \item Replace \code{sd_rm} by \code{\link{sd}} +#' \item Replace \code{var_rm} by \code{\link{var}} +#' \item Replace \code{sum_rm} by \code{\link{sum}} +#' \item Replace \code{getdata} by \code{\link{get_data}} +#' \item Replace \code{filterdata} by \code{\link{filter_data}} +#' \item Replace \code{combinedata} by \code{\link{combine_data}} +#' \item Replace \code{viewdata} by \code{\link{view_data}} +#' \item Replace \code{toFct} by \code{\link{to_fct}} +#' \item Replace \code{fixMS} by \code{\link{fix_smart}} +#' \item Replace \code{rounddf} by \code{\link{round_df}} +#' \item Replace \code{formatdf} by \code{\link{format_df}} +#' \item Replace \code{formatnr} by \code{\link{format_nr}} +#' \item Replace \code{getclass} by \code{\link{get_class}} +#' \item Replace \code{is_numeric} by \code{\link{is_double}} +#' \item Replace \code{is_empty} by \code{\link{is.empty}} +#' } +#' +mean_rm <- function(...) { + .Deprecated("mean") + mean(..., na.rm = TRUE) +} +median_rm <- function(...) { + .Deprecated("median") + median(..., na.rm = TRUE) +} +min_rm <- function(...) { + .Deprecated("min") + min(..., na.rm = TRUE) +} +max_rm <- function(...) { + .Deprecated("max") + max(..., na.rm = TRUE) +} +sd_rm <- function(...) { + .Deprecated("sd") + sd(..., na.rm = TRUE) +} +var_rm <- function(...) { + .Deprecated("var") + var(..., na.rm = TRUE) +} +sum_rm <- function(...) { + .Deprecated("sum") + sum(..., na.rm = TRUE) +} +getsummary <- function(...) { + .Deprecated("get_summary") + get_summary(...) +} +getdata <- function(...) { + .Deprecated("get_data") + get_data(...) +} +filterdata <- function(...) { + .Deprecated("filter_data") + filter_data(...) +} +combinedata <- function(...) { + .Deprecated("combine_data") + combine_data(...) +} +viewdata <- function(...) { + .Deprecated("view_data") + view_data(...) +} +toFct <- function(...) { + .Deprecated("to_fct") + to_fct(...) +} +fixMS <- function(...) { + .Deprecated("fix_smart") + fix_smart(...) +} +Search <- function(...) { + .Deprecated("search_data") + search_data(...) +} +formatnr <- function(...) { + .Deprecated("format_nr") + format_nr(...) +} +formatdf <- function(...) { + .Deprecated("format_df") + format_df(...) +} +rounddf <- function(...) { + .Deprecated("round_df") + round_df(...) +} +getclass <- function(...) { + .Deprecated("get_class") + get_class(...) +} +is_numeric <- function(...) { + .Deprecated("is_double") + is_double(...) +} +is_empty <- function(...) { + .Deprecated("is.empty") + is.empty(...) +} + +NULL diff --git a/radiant.data/R/explore.R b/radiant.data/R/explore.R new file mode 100644 index 0000000000000000000000000000000000000000..8d44fe9269be8b0b707ceaa0f806a6bca7d01416 --- /dev/null +++ b/radiant.data/R/explore.R @@ -0,0 +1,708 @@ +#' Explore and summarize data +#' +#' @details See \url{https://radiant-rstats.github.io/docs/data/explore.html} for an example in Radiant +#' +#' @param dataset Dataset to explore +#' @param vars (Numeric) variables to summarize +#' @param byvar Variable(s) to group data by +#' @param fun Functions to use for summarizing +#' @param top Use functions ("fun"), variables ("vars"), or group-by variables as column headers +#' @param tabfilt Expression used to filter the table (e.g., "Total > 10000") +#' @param tabsort Expression used to sort the table (e.g., "desc(Total)") +#' @param tabslice Expression used to filter table (e.g., "1:5") +#' @param nr Number of rows to display +#' @param data_filter Expression used to filter the dataset before creating the table (e.g., "price > 10000") +#' @param arr Expression to arrange (sort) the data on (e.g., "color, desc(price)") +#' @param rows Rows to select from the specified dataset +#' @param envir Environment to extract data from +#' +#' @return A list of all variables defined in the function as an object of class explore +#' +#' @examples +#' explore(diamonds, c("price", "carat")) %>% str() +#' explore(diamonds, "price:x")$tab +#' explore(diamonds, c("price", "carat"), byvar = "cut", fun = c("n_missing", "skew"))$tab +#' +#' @seealso See \code{\link{summary.explore}} to show summaries +#' +#' @export +explore <- function(dataset, vars = "", byvar = "", fun = c("mean", "sd"), + top = "fun", tabfilt = "", tabsort = "", tabslice = "", + nr = Inf, data_filter = "", arr = "", rows = NULL, + envir = parent.frame()) { + tvars <- vars + if (!is.empty(byvar)) tvars <- unique(c(tvars, byvar)) + + df_name <- if (is_string(dataset)) dataset else deparse(substitute(dataset)) + dataset <- get_data(dataset, tvars, filt = data_filter, arr = arr, rows = rows, na.rm = FALSE, envir = envir) + rm(tvars) + + ## in case : was used + vars <- base::setdiff(colnames(dataset), byvar) + + ## converting data as needed for summarization + dc <- get_class(dataset) + fixer <- function(x, fun = as_integer) { + if (is.character(x) || is.Date(x)) { + x <- rep(NA, length(x)) + } else if (is.factor(x)) { + x_num <- sshhr(as.integer(as.character(x))) + if (length(na.omit(x_num)) == 0) { + x <- fun(x) + } else { + x <- x_num + } + } + x + } + fixer_first <- function(x) { + x <- fixer(x, function(x) as_integer(x == levels(x)[1])) + } + mean <- function(x, na.rm = TRUE) sshhr(base::mean(fixer_first(x), na.rm = na.rm)) + sum <- function(x, na.rm = TRUE) sshhr(base::sum(fixer_first(x), na.rm = na.rm)) + var <- function(x, na.rm = TRUE) sshhr(stats::var(fixer_first(x), na.rm = na.rm)) + sd <- function(x, na.rm = TRUE) sshhr(stats::sd(fixer_first(x), na.rm = na.rm)) + se <- function(x, na.rm = TRUE) sshhr(radiant.data::se(fixer_first(x), na.rm = na.rm)) + me <- function(x, na.rm = TRUE) sshhr(radiant.data::me(fixer_first(x), na.rm = na.rm)) + cv <- function(x, na.rm = TRUE) sshhr(radiant.data::cv(fixer_first(x), na.rm = na.rm)) + prop <- function(x, na.rm = TRUE) sshhr(radiant.data::prop(fixer_first(x), na.rm = na.rm)) + varprop <- function(x, na.rm = TRUE) sshhr(radiant.data::varprop(fixer_first(x), na.rm = na.rm)) + sdprop <- function(x, na.rm = TRUE) sshhr(radiant.data::sdprop(fixer_first(x), na.rm = na.rm)) + seprop <- function(x, na.rm = TRUE) sshhr(radiant.data::seprop(fixer_first(x), na.rm = na.rm)) + meprop <- function(x, na.rm = TRUE) sshhr(radiant.data::meprop(fixer_first(x), na.rm = na.rm)) + varpop <- function(x, na.rm = TRUE) sshhr(radiant.data::varpop(fixer_first(x), na.rm = na.rm)) + sdpop <- function(x, na.rm = TRUE) sshhr(radiant.data::sdpop(fixer_first(x), na.rm = na.rm)) + + median <- function(x, na.rm = TRUE) sshhr(stats::median(fixer(x), na.rm = na.rm)) + min <- function(x, na.rm = TRUE) sshhr(base::min(fixer(x), na.rm = na.rm)) + max <- function(x, na.rm = TRUE) sshhr(base::max(fixer(x), na.rm = na.rm)) + p01 <- function(x, na.rm = TRUE) sshhr(radiant.data::p01(fixer(x), na.rm = na.rm)) + p025 <- function(x, na.rm = TRUE) sshhr(radiant.data::p025(fixer(x), na.rm = na.rm)) + p05 <- function(x, na.rm = TRUE) sshhr(radiant.data::p05(fixer(x), na.rm = na.rm)) + p10 <- function(x, na.rm = TRUE) sshhr(radiant.data::p10(fixer(x), na.rm = na.rm)) + p25 <- function(x, na.rm = TRUE) sshhr(radiant.data::p25(fixer(x), na.rm = na.rm)) + p75 <- function(x, na.rm = TRUE) sshhr(radiant.data::p75(fixer(x), na.rm = na.rm)) + p90 <- function(x, na.rm = TRUE) sshhr(radiant.data::p90(fixer(x), na.rm = na.rm)) + p95 <- function(x, na.rm = TRUE) sshhr(radiant.data::p95(fixer(x), na.rm = na.rm)) + p975 <- function(x, na.rm = TRUE) sshhr(radiant.data::p975(fixer(x), na.rm = na.rm)) + p99 <- function(x, na.rm = TRUE) sshhr(radiant.data::p99(fixer(x), na.rm = na.rm)) + skew <- function(x, na.rm = TRUE) sshhr(radiant.data::skew(fixer(x), na.rm = na.rm)) + kurtosi <- function(x, na.rm = TRUE) sshhr(radiant.data::kurtosi(fixer(x), na.rm = na.rm)) + + isLogNum <- "logical" == dc & names(dc) %in% base::setdiff(vars, byvar) + if (sum(isLogNum) > 0) { + dataset[, isLogNum] <- select(dataset, which(isLogNum)) %>% + mutate_all(as.integer) + dc[isLogNum] <- "integer" + } + + if (is.empty(byvar)) { + byvar <- c() + tab <- summarise_all(dataset, fun, na.rm = TRUE) + } else { + + ## convert categorical variables to factors if needed + ## needed to deal with empty/missing values + dataset[, byvar] <- select_at(dataset, .vars = byvar) %>% + mutate_all(~ empty_level(.)) + + tab <- dataset %>% + group_by_at(.vars = byvar) %>% + summarise_all(fun, na.rm = TRUE) + } + + ## adjust column names + if (length(vars) == 1 || length(fun) == 1) { + rng <- (length(byvar) + 1):ncol(tab) + colnames(tab)[rng] <- paste0(vars, "_", fun) + rm(rng) + } + + ## setup regular expression to split variable/function column appropriately + rex <- paste0("(.*?)_", glue('({glue_collapse(fun, "$|")}$)')) + + ## useful answer and comments: http://stackoverflow.com/a/27880388/1974918 + tab <- gather(tab, "variable", "value", !!-(seq_along(byvar))) %>% + extract(variable, into = c("variable", "fun"), regex = rex) %>% + mutate(fun = factor(fun, levels = !!fun), variable = factor(variable, levels = vars)) %>% + # mutate(variable = paste0(variable, " {", dc[variable], "}")) %>% + spread("fun", "value") + + ## flip the table if needed + if (top != "fun") { + tab <- list(tab = tab, byvar = byvar, fun = fun) %>% + flip(top) + } + + nrow_tab <- nrow(tab) + + ## filtering the table if desired from Report > Rmd + if (!is.empty(tabfilt)) { + tab <- filter_data(tab, tabfilt) + } + + ## sorting the table if desired from Report > Rmd + if (!identical(tabsort, "")) { + tabsort <- gsub(",", ";", tabsort) + tab <- tab %>% arrange(!!!rlang::parse_exprs(tabsort)) + } + + ## ensure factors ordered as in the (sorted) table + if (!is.empty(byvar) && top != "byvar") { + for (i in byvar) tab[[i]] <- tab[[i]] %>% (function(x) factor(x, levels = unique(x))) + rm(i) + } + + ## frequencies converted to doubles during gather/spread above + check_int <- function(x) { + if (is.double(x) && length(na.omit(x)) > 0) { + x_int <- sshhr(as.integer(round(x, .Machine$double.rounding))) + if (isTRUE(all.equal(x, x_int, check.attributes = FALSE))) x_int else x + } else { + x + } + } + + tab <- ungroup(tab) %>% mutate_all(check_int) + + ## slicing the table if desired + if (!is.empty(tabslice)) { + tab <- tab %>% + slice_data(tabslice) %>% + droplevels() + } + + ## convert to data.frame to maintain attributes + tab <- as.data.frame(tab, stringsAsFactors = FALSE) + attr(tab, "radiant_nrow") <- nrow_tab + if (!isTRUE(is.infinite(nr))) { + ind <- if (nr > nrow(tab)) 1:nrow(tab) else 1:nr + tab <- tab[ind, , drop = FALSE] + rm(ind) + } + + list( + tab = tab, + df_name = df_name, + vars = vars, + byvar = byvar, + fun = fun, + top = top, + tabfilt = tabfilt, + tabsort = tabsort, + tabslice = tabslice, + nr = nr, + data_filter = data_filter, + arr = arr, + rows = rows + ) %>% add_class("explore") +} + +#' Summary method for the explore function +#' +#' @details See \url{https://radiant-rstats.github.io/docs/data/explore.html} for an example in Radiant +#' +#' @param object Return value from \code{\link{explore}} +#' @param dec Number of decimals to show +#' @param ... further arguments passed to or from other methods +#' +#' @examples +#' result <- explore(diamonds, "price:x") +#' summary(result) +#' result <- explore(diamonds, "price", byvar = "cut", fun = c("n_obs", "skew")) +#' summary(result) +#' explore(diamonds, "price:x", byvar = "color") %>% summary() +#' +#' @seealso \code{\link{explore}} to generate summaries +#' +#' @export +summary.explore <- function(object, dec = 3, ...) { + cat("Explore\n") + cat("Data :", object$df_name, "\n") + if (!is.empty(object$data_filter)) { + cat("Filter :", gsub("\\n", "", object$data_filter), "\n") + } + if (!is.empty(object$arr)) { + cat("Arrange :", gsub("\\n", "", object$arr), "\n") + } + if (!is.empty(object$rows)) { + cat("Slice :", gsub("\\n", "", object$rows), "\n") + } + if (!is.empty(object$tabfilt)) { + cat("Table filter:", object$tabfilt, "\n") + } + if (!is.empty(object$tabsort[1])) { + cat("Table sorted:", paste0(object$tabsort, collapse = ", "), "\n") + } + if (!is.empty(object$tabslice)) { + cat("Table slice :", object$tabslice, "\n") + } + nr <- attr(object$tab, "radiant_nrow") + if (!isTRUE(is.infinite(nr)) && !isTRUE(is.infinite(object$nr)) && object$nr < nr) { + cat(paste0("Rows shown : ", object$nr, " (out of ", nr, ")\n")) + } + if (!is.empty(object$byvar[1])) { + cat("Grouped by :", object$byvar, "\n") + } + cat("Functions :", paste0(object$fun, collapse = ", "), "\n") + cat("Top :", c("fun" = "Function", "var" = "Variables", "byvar" = "Group by")[object$top], "\n") + cat("\n") + + format_df(object$tab, dec = dec, mark = ",") %>% + print(row.names = FALSE) + invisible() +} + +#' Deprecated: Store method for the explore function +#' +#' @details Return the summarized data. See \url{https://radiant-rstats.github.io/docs/data/explore.html} for an example in Radiant +#' +#' @param dataset Dataset +#' @param object Return value from \code{\link{explore}} +#' @param name Name to assign to the dataset +#' @param ... further arguments passed to or from other methods +#' +#' @seealso \code{\link{explore}} to generate summaries +#' +#' @export +store.explore <- function(dataset, object, name, ...) { + if (missing(name)) { + object$tab + } else { + stop( + paste0( + "This function is deprecated. Use the code below instead:\n\n", + name, " <- ", deparse(substitute(object)), "$tab\nregister(\"", + name, ")" + ), + call. = FALSE + ) + } +} + +#' Flip the DT table to put Function, Variable, or Group by on top +#' +#' @details See \url{https://radiant-rstats.github.io/docs/data/explore.html} for an example in Radiant +#' +#' @param expl Return value from \code{\link{explore}} +#' @param top The variable (type) to display at the top of the table ("fun" for Function, "var" for Variable, and "byvar" for Group by. "fun" is the default +#' +#' @examples +#' explore(diamonds, "price:x", top = "var") %>% summary() +#' explore(diamonds, "price", byvar = "cut", fun = c("n_obs", "skew"), top = "byvar") %>% summary() +#' +#' @seealso \code{\link{explore}} to calculate summaries +#' @seealso \code{\link{summary.explore}} to show summaries +#' @seealso \code{\link{dtab.explore}} to create the DT table +#' +#' @export +flip <- function(expl, top = "fun") { + cvars <- expl$byvar %>% + (function(x) if (is.empty(x[1])) character(0) else x) + if (top[1] == "var") { + expl$tab %<>% gather(".function", "value", !!-(1:(length(cvars) + 1))) %>% + spread("variable", "value") + expl$tab[[".function"]] %<>% factor(., levels = expl$fun) + } else if (top[1] == "byvar" && length(cvars) > 0) { + expl$tab %<>% gather(".function", "value", !!-(1:(length(cvars) + 1))) %>% + spread(!!cvars[1], "value") + expl$tab[[".function"]] %<>% factor(., levels = expl$fun) + + ## ensure we don't have invalid column names + colnames(expl$tab) <- fix_names(colnames(expl$tab)) + } + + expl$tab +} + +#' Make an interactive table of summary statistics +#' +#' @details See \url{https://radiant-rstats.github.io/docs/data/explore.html} for an example in Radiant +#' +#' @param object Return value from \code{\link{explore}} +#' @param dec Number of decimals to show +#' @param searchCols Column search and filter +#' @param order Column sorting +#' @param pageLength Page length +#' @param caption Table caption +#' @param ... further arguments passed to or from other methods +#' +#' @examples +#' \dontrun{ +#' tab <- explore(diamonds, "price:x") %>% dtab() +#' tab <- explore(diamonds, "price", byvar = "cut", fun = c("n_obs", "skew"), top = "byvar") %>% +#' dtab() +#' } +#' +#' @seealso \code{\link{pivotr}} to create a pivot table +#' @seealso \code{\link{summary.pivotr}} to show summaries +#' +#' @export +dtab.explore <- function(object, dec = 3, searchCols = NULL, + order = NULL, pageLength = NULL, + caption = NULL, ...) { + style <- if (exists("bslib_current_version") && "4" %in% bslib_current_version()) "bootstrap4" else "bootstrap" + tab <- object$tab + cn_all <- colnames(tab) + cn_num <- cn_all[sapply(tab, is.numeric)] + cn_cat <- cn_all[-which(cn_all %in% cn_num)] + isInt <- sapply(tab, is.integer) + isDbl <- sapply(tab, is_double) + dec <- ifelse(is.empty(dec) || dec < 0, 3, round(dec, 0)) + + top <- c("fun" = "Function", "var" = "Variables", "byvar" = paste0("Group by: ", object$byvar[1]))[object$top] + sketch <- shiny::withTags( + table( + thead( + tr( + th(" ", colspan = length(cn_cat)), + lapply(top, th, colspan = length(cn_num), class = "text-center") + ), + tr(lapply(cn_all, th)) + ) + ) + ) + + if (!is.empty(caption)) { + ## from https://github.com/rstudio/DT/issues/630#issuecomment-461191378 + caption <- shiny::tags$caption(style = "caption-side: bottom; text-align: left; font-size:100%;", caption) + } + + ## for display options see https://datatables.net/reference/option/dom + dom <- if (nrow(tab) < 11) "t" else "ltip" + fbox <- if (nrow(tab) > 5e6) "none" else list(position = "top") + dt_tab <- DT::datatable( + tab, + container = sketch, + caption = caption, + selection = "none", + rownames = FALSE, + filter = fbox, + ## must use fillContainer = FALSE to address + ## see https://github.com/rstudio/DT/issues/367 + ## https://github.com/rstudio/DT/issues/379 + fillContainer = FALSE, + style = style, + options = list( + dom = dom, + stateSave = TRUE, ## store state + searchCols = searchCols, + order = order, + columnDefs = list(list(orderSequence = c("desc", "asc"), targets = "_all")), + autoWidth = TRUE, + processing = FALSE, + pageLength = { + if (is.null(pageLength)) 10 else pageLength + }, + lengthMenu = list(c(5, 10, 25, 50, -1), c("5", "10", "25", "50", "All")) + ), + ## https://github.com/rstudio/DT/issues/146#issuecomment-534319155 + callback = DT::JS('$(window).on("unload", function() { table.state.clear(); })') + ) %>% + DT::formatStyle(., cn_cat, color = "white", backgroundColor = "grey") + + ## rounding as needed + if (sum(isDbl) > 0) { + dt_tab <- DT::formatRound(dt_tab, names(isDbl)[isDbl], dec) + } + if (sum(isInt) > 0) { + dt_tab <- DT::formatRound(dt_tab, names(isInt)[isInt], 0) + } + + ## see https://github.com/yihui/knitr/issues/1198 + dt_tab$dependencies <- c( + list(rmarkdown::html_dependency_bootstrap("bootstrap")), + dt_tab$dependencies + ) + + dt_tab +} + +########################################### +## turn functions below into functional ... +########################################### + +#' Number of observations +#' @param x Input variable +#' @param ... Additional arguments +#' @return number of observations +#' @examples +#' n_obs(c("a", "b", NA)) +#' +#' @export +n_obs <- function(x, ...) length(x) + +#' Number of missing values +#' @param x Input variable +#' @param ... Additional arguments +#' @return number of missing values +#' @examples +#' n_missing(c("a", "b", NA)) +#' +#' @export +n_missing <- function(x, ...) sum(is.na(x)) + +#' Calculate percentiles +#' @param x Numeric vector +#' @param na.rm If TRUE missing values are removed before calculation +#' @examples +#' p01(0:100) +#' +#' @rdname percentiles +#' @export +p01 <- function(x, na.rm = TRUE) quantile(x, .01, na.rm = na.rm) + +#' @rdname percentiles +#' @export +p025 <- function(x, na.rm = TRUE) quantile(x, .025, na.rm = na.rm) + +#' @rdname percentiles +#' @export +p05 <- function(x, na.rm = TRUE) quantile(x, .05, na.rm = na.rm) + +#' @rdname percentiles +#' @export +p10 <- function(x, na.rm = TRUE) quantile(x, .1, na.rm = na.rm) + +#' @rdname percentiles +#' @export +p25 <- function(x, na.rm = TRUE) quantile(x, .25, na.rm = na.rm) + +#' @rdname percentiles +#' @export +p75 <- function(x, na.rm = TRUE) quantile(x, .75, na.rm = na.rm) + +#' @rdname percentiles +#' @export +p90 <- function(x, na.rm = TRUE) quantile(x, .90, na.rm = na.rm) + +#' @rdname percentiles +#' @export +p95 <- function(x, na.rm = TRUE) quantile(x, .95, na.rm = na.rm) + +#' @rdname percentiles +#' @export +p975 <- function(x, na.rm = TRUE) quantile(x, .975, na.rm = na.rm) + +#' @rdname percentiles +#' @export +p99 <- function(x, na.rm = TRUE) quantile(x, .99, na.rm = na.rm) + +#' Coefficient of variation +#' @param x Input variable +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Coefficient of variation +#' @examples +#' cv(runif(100)) +#' +#' @export +cv <- function(x, na.rm = TRUE) { + m <- mean(x, na.rm = na.rm) + if (m == 0) { + message("Mean should be greater than 0") + NA + } else { + sd(x, na.rm = na.rm) / m + } +} + +#' Standard error +#' @param x Input variable +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Standard error +#' @examples +#' se(rnorm(100)) +#' +#' @export +se <- function(x, na.rm = TRUE) { + if (na.rm) x <- na.omit(x) + sd(x) / sqrt(length(x)) +} + +#' Margin of error +#' @param x Input variable +#' @param conf_lev Confidence level. The default is 0.95 +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Margin of error +#' +#' @importFrom stats qt +#' +#' @examples +#' me(rnorm(100)) +#' +#' @export +me <- function(x, conf_lev = 0.95, na.rm = TRUE) { + if (na.rm) x <- na.omit(x) + se(x) * qt(conf_lev / 2 + .5, length(x) - 1, lower.tail = TRUE) +} + +#' Calculate proportion +#' @param x Input variable +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Proportion of first level for a factor and of the maximum value for numeric +#' @examples +#' prop(c(rep(1L, 10), rep(0L, 10))) +#' prop(c(rep(4, 10), rep(2, 10))) +#' prop(rep(0, 10)) +#' prop(factor(c(rep("a", 20), rep("b", 10)))) +#' +#' @export +prop <- function(x, na.rm = TRUE) { + if (na.rm) x <- na.omit(x) + if (is.numeric(x)) { + mean(x == max(x, 1)) ## gives proportion of max value in x + } else if (is.factor(x)) { + mean(x == levels(x)[1]) ## gives proportion of first level in x + } else if (is.logical(x)) { + mean(x) + } else { + NA + } +} + +#' Variance for proportion +#' @param x Input variable +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Variance for proportion +#' @examples +#' varprop(c(rep(1L, 10), rep(0L, 10))) +#' +#' @export +varprop <- function(x, na.rm = TRUE) { + p <- prop(x, na.rm = na.rm) + p * (1 - p) +} + +#' Standard deviation for proportion +#' @param x Input variable +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Standard deviation for proportion +#' @examples +#' sdprop(c(rep(1L, 10), rep(0L, 10))) +#' +#' @export +sdprop <- function(x, na.rm = TRUE) sqrt(varprop(x, na.rm = na.rm)) + +#' Standard error for proportion +#' @param x Input variable +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Standard error for proportion +#' @examples +#' seprop(c(rep(1L, 10), rep(0L, 10))) +#' +#' @export +seprop <- function(x, na.rm = TRUE) { + if (na.rm) x <- na.omit(x) + sqrt(varprop(x, na.rm = FALSE) / length(x)) +} + +#' Margin of error for proportion +#' @param x Input variable +#' @param conf_lev Confidence level. The default is 0.95 +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Margin of error +#' +#' @importFrom stats qnorm +#' +#' @examples +#' meprop(c(rep(1L, 10), rep(0L, 10))) +#' +#' @export +meprop <- function(x, conf_lev = 0.95, na.rm = TRUE) { + if (na.rm) x <- na.omit(x) + seprop(x) * qnorm(conf_lev / 2 + .5, lower.tail = TRUE) +} + +#' Variance for the population +#' @param x Input variable +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Variance for the population +#' @examples +#' varpop(rnorm(100)) +#' +#' @export +varpop <- function(x, na.rm = TRUE) { + if (na.rm) x <- na.omit(x) + n <- length(x) + var(x) * ((n - 1) / n) +} + +#' Standard deviation for the population +#' @param x Input variable +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Standard deviation for the population +#' @examples +#' sdpop(rnorm(100)) +#' +#' @export +sdpop <- function(x, na.rm = TRUE) sqrt(varpop(x, na.rm = na.rm)) + +#' Natural log +#' @param x Input variable +#' @param na.rm Remove missing values (default is TRUE) +#' @return Natural log of vector +#' @examples +#' ln(runif(10, 1, 2)) +#' +#' @export +ln <- function(x, na.rm = TRUE) { + if (na.rm) log(na.omit(x)) else log(x) +} + +#' Does a vector have non-zero variability? +#' @param x Input variable +#' @param na.rm If TRUE missing values are removed before calculation +#' @return Logical. TRUE is there is variability +#' @examples +#' summarise_all(diamonds, does_vary) %>% as.logical() +#' +#' @export +does_vary <- function(x, na.rm = TRUE) { + ## based on http://stackoverflow.com/questions/4752275/test-for-equality-among-all-elements-of-a-single-vector + if (length(x) == 1L) { + FALSE + } else { + if (is.factor(x) || is.character(x)) { + length(unique(x)) > 1 + } else { + abs(max(x, na.rm = na.rm) - min(x, na.rm = na.rm)) > .Machine$double.eps^0.5 + } + } +} + +#' Convert categorical variables to factors and deal with empty/missing values +#' @param x Categorical variable used in table +#' @return Variable with updated levels +#' @export +empty_level <- function(x) { + if (!is.factor(x)) x <- as.factor(x) + levs <- levels(x) + if ("" %in% levs) { + levs[levs == ""] <- "NA" + x <- factor(x, levels = levs) + x[is.na(x)] <- "NA" + } else if (any(is.na(x))) { + x <- factor(x, levels = unique(c(levs, "NA"))) + x[is.na(x)] <- "NA" + } + x +} + +#' Calculate the mode (modal value) and return a label +#' +#' @details From https://www.tutorialspoint.com/r/r_mean_median_mode.htm +#' @param x A vector +#' @param na.rm If TRUE missing values are removed before calculation +#' +#' @examples +#' modal(c("a", "b", "b")) +#' modal(c(1:10, 5)) +#' modal(as.factor(c(letters, "b"))) +#' modal(runif(100) > 0.5) +#' +#' @export +modal <- function(x, na.rm = TRUE) { + if (na.rm) x <- na.omit(x) + unv <- unique(x) + unv[which.max(tabulate(match(x, unv)))] +} diff --git a/radiant.data/R/for.shinyapps.io.R b/radiant.data/R/for.shinyapps.io.R new file mode 100644 index 0000000000000000000000000000000000000000..14ab46e6a4edffc93a646bd2fddacc714aad6133 --- /dev/null +++ b/radiant.data/R/for.shinyapps.io.R @@ -0,0 +1,29 @@ +# ## install the latest version from github so it will be used on shinyapps.io +# packages <- "radiant-rstats/radiant.data" +# packages <- c(packages, "trestletech/shinyAce", "thomasp85/shinyFiles") +# +# ## Use the code below to install the development version +# if (!require(remotes)) { +# install.packages("remotes") +# } +# ret <- sapply( +# packages, +# function(p) { +# remotes::install_github( +# p, +# dependencies = FALSE, +# upgrade = "never" +# ) +# } +# ) +# +# # install.packages("htmltools", repo = "https://cloud.r-project.org/") +# +# ## by listing the call to the radiant library it will get picked up as a dependency +# library(radiant) +# library(radiant.data) +# library(rstudioapi) +# library(shinyAce) +# library(shinyFiles) +# library(DT) +# library(htmltools) diff --git a/radiant.data/R/manage.R b/radiant.data/R/manage.R new file mode 100644 index 0000000000000000000000000000000000000000..e7fdd79f52192b8a416345b08ff776ba52768db9 --- /dev/null +++ b/radiant.data/R/manage.R @@ -0,0 +1,88 @@ +#' Load data through clipboard on Windows or macOS +#' +#' @details Extract data from the clipboard into a data.frame on Windows or macOS +#' @param delim Delimiter to use (tab is the default) +#' @param text Text input to convert to table +#' @param suppress Suppress warnings +#' @seealso See the \code{\link{save_clip}} +#' @export +load_clip <- function(delim = "\t", text, suppress = TRUE) { + sw <- if (suppress) suppressWarnings else function(x) x + sw( + try( + { + os_type <- Sys.info()["sysname"] + if (os_type == "Windows") { + dataset <- read.table( + "clipboard", + header = TRUE, sep = delim, + comment.char = "", fill = TRUE, as.is = TRUE, + check.names = FALSE + ) + } else if (os_type == "Darwin") { + dataset <- read.table( + pipe("pbpaste"), + header = TRUE, sep = delim, + comment.char = "", fill = TRUE, as.is = TRUE, + check.names = FALSE + ) + } else if (os_type == "Linux") { + if (missing(text) || is.empty(text)) { + message("Loading data through clipboard is currently only supported on Windows and macOS") + return(invisible()) + } else { + dataset <- read.table( + text = text, header = TRUE, sep = delim, + comment.char = "", fill = TRUE, as.is = TRUE, + check.names = FALSE + ) + } + } + as.data.frame(dataset, check.names = FALSE, stringsAsFactors = FALSE) %>% + radiant.data::to_fct() + }, + silent = TRUE + ) + ) +} + +#' Save data to clipboard on Windows or macOS +#' +#' @details Save a data.frame or tibble to the clipboard on Windows or macOS +#' @param dataset Dataset to save to clipboard +#' @seealso See the \code{\link{load_clip}} +#' @export +save_clip <- function(dataset) { + os_type <- Sys.info()["sysname"] + if (os_type == "Windows") { + write.table(dataset, "clipboard-10000", sep = "\t", row.names = FALSE) + } else if (os_type == "Darwin") { + write.table(dataset, file = pipe("pbcopy"), sep = "\t", row.names = FALSE) + } else if (os_type == "Linux") { + message("Saving data to clipboard is currently only supported on Windows and macOS.\nSave data to csv for use in a spreadsheet") + } + invisible() +} + +#' Ensure column names are valid +#' +#' @details Remove symbols, trailing and leading spaces, and convert to valid R column names. Opinionated version of \code{\link{make.names}} +#' @param x Data.frame or vector of (column) names +#' @param lower Set letters to lower case (TRUE or FALSE) +#' @examples +#' fix_names(c(" var-name ", "$amount spent", "100")) +#' @export +fix_names <- function(x, lower = FALSE) { + isdf <- is.data.frame(x) + cn <- if (isdf) colnames(x) else x + cn <- gsub("(^\\s+|\\s+$)", "", cn) %>% + gsub("\\s+", "_", .) %>% + gsub("[[:punct:]]", "_", .) %>% + gsub("^[[:punct:]]", "", .) %>% + make.names(unique = TRUE) %>% + gsub("\\.{2,}", ".", .) %>% + gsub("_{2,}", "_", .) %>% + make.names(unique = TRUE) %>% ## used twice to make sure names are still unique + (function(x) if (lower) tolower(x) else x) + if (isdf) stats::setNames(x, cn) else cn +} \ No newline at end of file diff --git a/radiant.data/R/pivotr.R b/radiant.data/R/pivotr.R new file mode 100644 index 0000000000000000000000000000000000000000..9103605b0faa6d21554a22e395489db9d62f99a5 --- /dev/null +++ b/radiant.data/R/pivotr.R @@ -0,0 +1,572 @@ +#' Create a pivot table +#' +#' @details Create a pivot-table. See \url{https://radiant-rstats.github.io/docs/data/pivotr.html} for an example in Radiant +#' +#' @param dataset Dataset to tabulate +#' @param cvars Categorical variables +#' @param nvar Numerical variable +#' @param fun Function to apply to numerical variable +#' @param normalize Normalize the table by row total, column totals, or overall total +#' @param tabfilt Expression used to filter the table (e.g., "Total > 10000") +#' @param tabsort Expression used to sort the table (e.g., "desc(Total)") +#' @param tabslice Expression used to filter table (e.g., "1:5") +#' @param nr Number of rows to display +#' @param data_filter Expression used to filter the dataset before creating the table (e.g., "price > 10000") +#' @param arr Expression to arrange (sort) the data on (e.g., "color, desc(price)") +#' @param rows Rows to select from the specified dataset +#' @param envir Environment to extract data from +#' +#' @examples +#' pivotr(diamonds, cvars = "cut") %>% str() +#' pivotr(diamonds, cvars = "cut")$tab +#' pivotr(diamonds, cvars = c("cut", "clarity", "color"))$tab +#' pivotr(diamonds, cvars = "cut:clarity", nvar = "price")$tab +#' pivotr(diamonds, cvars = "cut", nvar = "price")$tab +#' pivotr(diamonds, cvars = "cut", normalize = "total")$tab +#' +#' @export +pivotr <- function(dataset, cvars = "", nvar = "None", fun = "mean", + normalize = "None", tabfilt = "", tabsort = "", tabslice = "", + nr = Inf, data_filter = "", arr = "", rows = NULL, envir = parent.frame()) { + vars <- if (nvar == "None") cvars else c(cvars, nvar) + fill <- if (nvar == "None") 0L else NA + + df_name <- if (is_string(dataset)) dataset else deparse(substitute(dataset)) + dataset <- get_data(dataset, vars, filt = data_filter, arr = arr, rows = rows, na.rm = FALSE, envir = envir) + + ## in case : was used for cvars + cvars <- base::setdiff(colnames(dataset), nvar) + + if (nvar == "None") { + nvar <- "n_obs" + } else { + fixer <- function(x, fun = as_integer) { + if (is.character(x) || is.Date(x)) { + x <- rep(NA, length(x)) + } else if (is.factor(x)) { + x_num <- sshhr(as.integer(as.character(x))) + if (length(na.omit(x_num)) == 0) { + x <- fun(x) + } else { + x <- x_num + } + } + x + } + fixer_first <- function(x) { + x <- fixer(x, function(x) as_integer(x == levels(x)[1])) + } + if (fun %in% c("mean", "sum", "sd", "var", "sd", "se", "me", "cv", "prop", "varprop", "sdprop", "seprop", "meprop", "varpop", "sepop")) { + dataset[[nvar]] <- fixer_first(dataset[[nvar]]) + } else if (fun %in% c("median", "min", "max", "p01", "p025", "p05", "p10", "p25", "p50", "p75", "p90", "p95", "p975", "p99", "skew", "kurtosi")) { + dataset[[nvar]] <- fixer(dataset[[nvar]]) + } + rm(fixer, fixer_first) + if ("logical" %in% class(dataset[[nvar]])) { + dataset[[nvar]] %<>% as.integer() + } + } + + ## convert categorical variables to factors and deal with empty/missing values + dataset <- mutate_at(dataset, .vars = cvars, .funs = empty_level) + + sel <- function(x, nvar, cvar = c()) { + if (nvar == "n_obs") x else select_at(x, .vars = c(nvar, cvar)) + } + sfun <- function(x, nvar, cvars = "", fun = fun) { + if (nvar == "n_obs") { + if (is.empty(cvars)) { + count(x) %>% dplyr::rename("n_obs" = "n") + } else { + count(select_at(x, .vars = cvars)) %>% dplyr::rename("n_obs" = "n") + } + } else { + dataset <- mutate_at(x, .vars = nvar, .funs = as.numeric) %>% + summarise_at(.vars = nvar, .funs = fun, na.rm = TRUE) + colnames(dataset)[ncol(dataset)] <- nvar + dataset + } + } + + ## main tab + tab <- dataset %>% + group_by_at(.vars = cvars) %>% + sfun(nvar, cvars, fun) + + ## total + total <- dataset %>% + sel(nvar) %>% + sfun(nvar, fun = fun) + + ## row and column totals + if (length(cvars) == 1) { + tab <- + bind_rows( + mutate_at(ungroup(tab), .vars = cvars, .funs = as.character), + bind_cols( + data.frame("Total", stringsAsFactors = FALSE) %>% + setNames(cvars), total %>% + set_colnames(nvar) + ) + ) + } else { + col_total <- + group_by_at(dataset, .vars = cvars[1]) %>% + sel(nvar, cvars[1]) %>% + sfun(nvar, cvars[1], fun) %>% + ungroup() %>% + mutate_at(.vars = cvars[1], .funs = as.character) + + row_total <- + group_by_at(dataset, .vars = cvars[-1]) %>% + sfun(nvar, cvars[-1], fun) %>% + ungroup() %>% + select(ncol(.)) %>% + bind_rows(total) %>% + set_colnames("Total") + + ## creating cross tab + tab <- spread(tab, !!cvars[1], !!nvar, fill = fill) %>% + ungroup() %>% + mutate_at(.vars = cvars[-1], .funs = as.character) + + tab <- bind_rows( + tab, + bind_cols( + t(rep("Total", length(cvars[-1]))) %>% + as.data.frame(stringsAsFactors = FALSE) %>% + setNames(cvars[-1]), + data.frame(t(col_total[[2]]), stringsAsFactors = FALSE) %>% + set_colnames(col_total[[1]]) + ) + ) %>% bind_cols(row_total) + + rm(col_total, row_total, vars) + } + + ## resetting factor levels + ind <- ifelse(length(cvars) > 1, -1, 1) + levs <- lapply(select_at(dataset, .vars = cvars[ind]), levels) + + for (i in cvars[ind]) { + tab[[i]] %<>% factor(levels = unique(c(levs[[i]], "Total"))) + } + + ## frequency table for chi-square test + tab_freq <- tab + + isNum <- if (length(cvars) == 1) -1 else -c(1:(length(cvars) - 1)) + if (normalize == "total") { + tab[, isNum] %<>% (function(x) x / total[[1]]) + } else if (normalize == "row") { + if (!is.null(tab[["Total"]])) { + tab[, isNum] %<>% (function(x) x / x[["Total"]]) + } + } else if (length(cvars) > 1 && normalize == "column") { + tab[, isNum] %<>% apply(2, function(.) . / .[which(tab[, 1] == "Total")]) + } + + nrow_tab <- nrow(tab) - 1 + + ## ensure we don't have invalid column names + ## but skip variable names already being used + cn <- colnames(tab) + cni <- cn %in% setdiff(cn, c(cvars, nvar)) + colnames(tab)[cni] <- fix_names(cn[cni]) + + ## filtering the table if desired + if (!is.empty(tabfilt)) { + tab <- tab[-nrow(tab), ] %>% + filter_data(tabfilt, drop = FALSE) %>% + bind_rows(tab[nrow(tab), ]) %>% + droplevels() + } + + ## sorting the table if desired + if (!is.empty(tabsort, "")) { + tabsort <- gsub(",", ";", tabsort) + tab[-nrow(tab), ] %<>% arrange(!!!rlang::parse_exprs(tabsort)) + + ## order factors as set in the sorted table + tc <- if (length(cvars) == 1) cvars else cvars[-1] ## don't change top cv + for (i in tc) { + tab[[i]] %<>% factor(., levels = unique(.)) + } + } + + ## slicing the table if desired + if (!is.empty(tabslice)) { + tab <- tab %>% + slice_data(tabslice) %>% + bind_rows(tab[nrow(tab), , drop = FALSE]) %>% + droplevels() + } + + tab <- as.data.frame(tab, stringsAsFactors = FALSE) + attr(tab, "radiant_nrow") <- nrow_tab + if (!isTRUE(is.infinite(nr))) { + ind <- if (nr >= nrow(tab)) 1:nrow(tab) else c(1:nr, nrow(tab)) + tab <- tab[ind, , drop = FALSE] + } + + rm(isNum, dataset, sfun, sel, i, levs, total, ind, nrow_tab, envir) + + as.list(environment()) %>% add_class("pivotr") +} + +#' Summary method for pivotr +#' +#' @details See \url{https://radiant-rstats.github.io/docs/data/pivotr.html} for an example in Radiant +#' +#' @param object Return value from \code{\link{pivotr}} +#' @param perc Display numbers as percentages (TRUE or FALSE) +#' @param dec Number of decimals to show +#' @param chi2 If TRUE calculate the chi-square statistic for the (pivot) table +#' @param shiny Did the function call originate inside a shiny app +#' @param ... further arguments passed to or from other methods +#' +#' @examples +#' pivotr(diamonds, cvars = "cut") %>% summary(chi2 = TRUE) +#' pivotr(diamonds, cvars = "cut", tabsort = "desc(n_obs)") %>% summary() +#' pivotr(diamonds, cvars = "cut", tabfilt = "n_obs > 700") %>% summary() +#' pivotr(diamonds, cvars = "cut:clarity", nvar = "price") %>% summary() +#' +#' @seealso \code{\link{pivotr}} to create the pivot-table using dplyr +#' +#' @export +summary.pivotr <- function(object, perc = FALSE, dec = 3, + chi2 = FALSE, shiny = FALSE, ...) { + if (!shiny) { + cat("Pivot table\n") + cat("Data :", object$df_name, "\n") + if (!is.empty(object$data_filter)) { + cat("Filter :", gsub("\\n", "", object$data_filter), "\n") + } + if (!is.empty(object$arr)) { + cat("Arrange :", gsub("\\n", "", object$arr), "\n") + } + if (!is.empty(object$rows)) { + cat("Slice :", gsub("\\n", "", object$rows), "\n") + } + if (!is.empty(object$tabfilt)) { + cat("Table filter:", object$tabfilt, "\n") + } + if (!is.empty(object$tabsort[1])) { + cat("Table sorted:", paste0(object$tabsort, collapse = ", "), "\n") + } + if (!is.empty(object$tabslice)) { + cat("Table slice :", object$tabslice, "\n") + } + nr <- attr(object$tab, "radiant_nrow") + if (!isTRUE(is.infinite(nr)) && !isTRUE(is.infinite(object$nr)) && object$nr < nr) { + cat(paste0("Rows shown : ", object$nr, " (out of ", nr, ")\n")) + } + cat("Categorical :", object$cvars, "\n") + if (object$normalize != "None") { + cat("Normalize by:", object$normalize, "\n") + } + if (object$nvar != "n_obs") { + cat("Numeric :", object$nvar, "\n") + cat("Function :", object$fun, "\n") + } + cat("\n") + print(format_df(object$tab, dec, perc, mark = ","), row.names = FALSE) + cat("\n") + } + + if (chi2) { + if (length(object$cvars) < 3) { + cst <- object$tab_freq %>% + filter(.[[1]] != "Total") %>% + select(-which(names(.) %in% c(object$cvars, "Total"))) %>% + mutate_all(~ ifelse(is.na(.), 0, .)) %>% + { + sshhr(chisq.test(., correct = FALSE)) + } + + res <- tidy(cst) + if (dec < 4 && res$p.value < .001) { + p.value <- "< .001" + } else { + p.value <- format_nr(res$p.value, dec = dec) + } + res <- round_df(res, dec) + + l1 <- paste0("Chi-squared: ", res$statistic, " df(", res$parameter, "), p.value ", p.value, "\n") + l2 <- paste0(sprintf("%.1f", 100 * (sum(cst$expected < 5) / length(cst$expected))), "% of cells have expected values below 5\n") + if (nrow(object$tab_freq) == nrow(object$tab)) { + if (shiny) HTML(paste0("
", l1, "", l2)) else cat(paste0(l1, l2)) + } else { + note <- "\nNote: Test conducted on unfiltered table" + if (shiny) HTML(paste0("
", l1, "", l2, "
", note)) else cat(paste0(l1, l2, note)) + } + } else { + cat("The number of categorical variables should be 1 or 2 for Chi-square") + } + } +} + +#' Make an interactive pivot table +#' +#' @details See \url{https://radiant-rstats.github.io/docs/data/pivotr.html} for an example in Radiant +#' +#' @param object Return value from \code{\link{pivotr}} +#' @param format Show Color bar ("color_bar"), Heat map ("heat"), or None ("none") +#' @param perc Display numbers as percentages (TRUE or FALSE) +#' @param dec Number of decimals to show +#' @param searchCols Column search and filter +#' @param order Column sorting +#' @param pageLength Page length +#' @param caption Table caption +#' @param ... further arguments passed to or from other methods +#' +#' @examples +#' \dontrun{ +#' pivotr(diamonds, cvars = "cut") %>% dtab() +#' pivotr(diamonds, cvars = c("cut", "clarity")) %>% dtab(format = "color_bar") +#' pivotr(diamonds, cvars = c("cut", "clarity"), normalize = "total") %>% +#' dtab(format = "color_bar", perc = TRUE) +#' } +#' +#' @seealso \code{\link{pivotr}} to create the pivot table +#' @seealso \code{\link{summary.pivotr}} to print the table +#' +#' @export +dtab.pivotr <- function(object, format = "none", perc = FALSE, dec = 3, + searchCols = NULL, order = NULL, pageLength = NULL, + caption = NULL, ...) { + style <- if (exists("bslib_current_version") && "4" %in% bslib_current_version()) "bootstrap4" else "bootstrap" + tab <- object$tab + cvar <- object$cvars[1] + cvars <- object$cvars %>% + (function(x) if (length(x) > 1) x[-1] else x) + cn <- colnames(tab) %>% + (function(x) x[-which(cvars %in% x)]) + + ## for rounding + isDbl <- sapply(tab, is_double) + isInt <- sapply(tab, is.integer) + dec <- ifelse(is.empty(dec) || dec < 0, 3, round(dec, 0)) + + ## column names without total + cn_nt <- if ("Total" %in% cn) cn[-which(cn == "Total")] else cn + + tot <- tail(tab, 1)[-(1:length(cvars))] %>% + format_df(perc = perc, dec = dec, mark = ",") + + if (length(cvars) == 1 && cvar == cvars) { + sketch <- shiny::withTags(table( + thead(tr(lapply(c(cvars, cn), th))), + tfoot(tr(lapply(c("Total", tot), th))) + )) + } else { + sketch <- shiny::withTags(table( + thead( + tr(th(colspan = length(c(cvars, cn)), cvar, class = "dt-center")), + tr(lapply(c(cvars, cn), th)) + ), + tfoot( + tr(th(colspan = length(cvars), "Total"), lapply(tot, th)) + ) + )) + } + + if (!is.empty(caption)) { + ## from https://github.com/rstudio/DT/issues/630#issuecomment-461191378 + caption <- shiny::tags$caption(style = "caption-side: bottom; text-align: left; font-size:100%;", caption) + } + + + ## remove row with column totals + ## should perhaps be part of pivotr but convenient for now in tfoot + ## and for external calls to pivotr + tab <- filter(tab, tab[[1]] != "Total") + ## for display options see https://datatables.net/reference/option/dom + dom <- if (nrow(tab) < 11) "t" else "ltip" + fbox <- if (nrow(tab) > 5e6) "none" else list(position = "top") + dt_tab <- DT::datatable( + tab, + container = sketch, + caption = caption, + selection = "none", + rownames = FALSE, + filter = fbox, + ## must use fillContainer = FALSE to address + ## see https://github.com/rstudio/DT/issues/367 + ## https://github.com/rstudio/DT/issues/379 + fillContainer = FALSE, + style = style, + options = list( + dom = dom, + stateSave = TRUE, ## store state + searchCols = searchCols, + order = order, + columnDefs = list(list(orderSequence = c("desc", "asc"), targets = "_all")), + autoWidth = TRUE, + processing = FALSE, + pageLength = { + if (is.null(pageLength)) 10 else pageLength + }, + lengthMenu = list(c(5, 10, 25, 50, -1), c("5", "10", "25", "50", "All")) + ), + ## https://github.com/rstudio/DT/issues/146#issuecomment-534319155 + callback = DT::JS('$(window).on("unload", function() { table.state.clear(); })') + ) %>% + DT::formatStyle(., cvars, color = "white", backgroundColor = "grey") %>% + (function(x) if ("Total" %in% cn) DT::formatStyle(x, "Total", fontWeight = "bold") else x) + + ## heat map with red or color_bar + if (format == "color_bar") { + dt_tab <- DT::formatStyle( + dt_tab, + cn_nt, + background = DT::styleColorBar(range(tab[, cn_nt], na.rm = TRUE), "lightblue"), + backgroundSize = "98% 88%", + backgroundRepeat = "no-repeat", + backgroundPosition = "center" + ) + } else if (format == "heat") { + ## round seems to ensure that 'cuts' are ordered according to DT::stylInterval + brks <- quantile(tab[, cn_nt], probs = seq(.05, .95, .05), na.rm = TRUE) %>% round(5) + clrs <- seq(255, 40, length.out = length(brks) + 1) %>% + round(0) %>% + (function(x) paste0("rgb(255,", x, ",", x, ")")) + + dt_tab <- DT::formatStyle(dt_tab, cn_nt, backgroundColor = DT::styleInterval(brks, clrs)) + } + + if (perc) { + ## show percentages + dt_tab <- DT::formatPercentage(dt_tab, cn, dec) + } else { + if (sum(isDbl) > 0) { + dt_tab <- DT::formatRound(dt_tab, names(isDbl)[isDbl], dec) + } + if (sum(isInt) > 0) { + dt_tab <- DT::formatRound(dt_tab, names(isInt)[isInt], 0) + } + } + + ## see https://github.com/yihui/knitr/issues/1198 + dt_tab$dependencies <- c( + list(rmarkdown::html_dependency_bootstrap("bootstrap")), + dt_tab$dependencies + ) + + dt_tab +} + +#' Plot method for the pivotr function +#' +#' @details See \url{https://radiant-rstats.github.io/docs/data/pivotr} for an example in Radiant +#' +#' @param x Return value from \code{\link{pivotr}} +#' @param type Plot type to use ("fill" or "dodge" (default)) +#' @param perc Use percentage on the y-axis +#' @param flip Flip the axes in a plot (FALSE or TRUE) +#' @param fillcol Fill color for bar-plot when only one categorical variable has been selected (default is "blue") +#' @param opacity Opacity for plot elements (0 to 1) +#' @param ... further arguments passed to or from other methods +#' +#' @examples +#' pivotr(diamonds, cvars = "cut") %>% plot() +#' pivotr(diamonds, cvars = c("cut", "clarity")) %>% plot() +#' pivotr(diamonds, cvars = c("cut", "clarity", "color")) %>% plot() +#' +#' @seealso \code{\link{pivotr}} to generate summaries +#' @seealso \code{\link{summary.pivotr}} to show summaries +#' +#' @importFrom rlang .data +#' +#' @export +plot.pivotr <- function(x, type = "dodge", perc = FALSE, flip = FALSE, + fillcol = "blue", opacity = 0.5, ...) { + cvars <- x$cvars + nvar <- x$nvar + tab <- x$tab %>% + (function(x) filter(x, x[[1]] != "Total")) + + if (flip) { + # need reverse order here because of how coord_flip works + tab <- lapply(tab, function(x) if (inherits(x, "factor")) factor(x, levels = rev(levels(x))) else x) %>% + as_tibble() + } + + if (length(cvars) == 1) { + p <- ggplot(na.omit(tab), aes(x = .data[[cvars]], y = .data[[nvar]])) + + geom_bar(stat = "identity", position = "dodge", alpha = opacity, fill = fillcol) + } else if (length(cvars) == 2) { + ctot <- which(colnames(tab) == "Total") + if (length(ctot) > 0) tab %<>% select(base::setdiff(colnames(.), "Total")) + + dots <- paste0("factor(", cvars[1], ", levels = c('", paste0(base::setdiff(colnames(tab), cvars[2]), collapse = "','"), "'))") %>% + rlang::parse_exprs(.) %>% + set_names(cvars[1]) + + p <- tab %>% + gather(!!cvars[1], !!nvar, !!base::setdiff(colnames(.), cvars[2])) %>% + na.omit() %>% + mutate(!!!dots) %>% + ggplot(aes(x = .data[[cvars[1]]], y = .data[[nvar]], fill = .data[[cvars[2]]])) + + geom_bar(stat = "identity", position = type, alpha = opacity) + } else if (length(cvars) == 3) { + ctot <- which(colnames(tab) == "Total") + if (length(ctot) > 0) tab %<>% select(base::setdiff(colnames(.), "Total")) + + dots <- paste0("factor(", cvars[1], ", levels = c('", paste0(base::setdiff(colnames(tab), cvars[2:3]), collapse = "','"), "'))") %>% + rlang::parse_exprs(.) %>% + set_names(cvars[1]) + + p <- tab %>% + gather(!!cvars[1], !!nvar, !!base::setdiff(colnames(.), cvars[2:3])) %>% + na.omit() %>% + mutate(!!!dots) %>% + ggplot(aes(x = .data[[cvars[1]]], y = .data[[nvar]], fill = .data[[cvars[2]]])) + + geom_bar(stat = "identity", position = type, alpha = opacity) + + facet_grid(paste(cvars[3], "~ .")) + } else { + ## No plot returned if more than 3 grouping variables are selected + return(invisible()) + } + + if (flip) p <- p + coord_flip() + if (perc) p <- p + scale_y_continuous(labels = scales::percent) + + if (isTRUE(nvar == "n_obs")) { + if (!is.empty(x$normalize, "None")) { + p <- p + labs(y = ifelse(perc, "Percentage", "Proportion")) + } + } else { + p <- p + labs(y = paste0(nvar, " (", x$fun, ")")) + } + + sshhr(p) +} + + +#' Deprecated: Store method for the pivotr function +#' +#' @details Return the summarized data. See \url{https://radiant-rstats.github.io/docs/data/pivotr.html} for an example in Radiant +#' +#' @param dataset Dataset +#' @param object Return value from \code{\link{pivotr}} +#' @param name Name to assign to the dataset +#' @param ... further arguments passed to or from other methods +#' +#' @seealso \code{\link{pivotr}} to generate summaries +#' +#' @export +store.pivotr <- function(dataset, object, name, ...) { + if (missing(name)) { + object$tab + } else { + stop( + paste0( + "This function is deprecated. Use the code below instead:\n\n", + name, " <- ", deparse(substitute(object)), "$tab\nregister(\"", + name, ")" + ), + call. = FALSE + ) + } +} diff --git a/radiant.data/R/radiant.R b/radiant.data/R/radiant.R new file mode 100644 index 0000000000000000000000000000000000000000..f15d919998bd06b2fdd8fc8a362c8e1c10bcfe54 --- /dev/null +++ b/radiant.data/R/radiant.R @@ -0,0 +1,1617 @@ +#' Launch radiant apps +#' +#' @details See \url{https://radiant-rstats.github.io/docs/} for radiant documentation and tutorials +#' +#' @param package Radiant package to start. One of "radiant.data", "radiant.design", "radiant.basics", "radiant.model", "radiant.multivariate", or "radiant" +#' @param run Run a radiant app in an external browser ("browser"), an Rstudio window ("window"), or in the Rstudio viewer ("viewer") +#' @param state Path to statefile to load +#' @param ... additional arguments to pass to shiny::runApp (e.g, port = 8080) +#' +#' @importFrom shiny paneViewer +#' +#' @examples +#' \dontrun{ +#' launch() +#' launch(run = "viewer") +#' launch(run = "window") +#' launch(run = "browser") +#' } +#' +#' @export +launch <- function(package = "radiant.data", run = "viewer", state, ...) { + ## check if package attached + if (!paste0("package:", package) %in% search()) { + if (!suppressWarnings(suppressMessages(suppressPackageStartupMessages(require(package, character.only = TRUE))))) { + stop(sprintf("Calling %s start function but %s is not installed.", package, package)) + } + } + + ## from Yihui's DT::datatable function + oop <- base::options( + width = max(getOption("width", 250), 250), + scipen = max(getOption("scipen", 100), 100), + max.print = max(getOption("max.print", 5000), 5000), + stringsAsFactors = FALSE, + radiant.launch_dir = normalizePath(getwd(), winslash = "/"), + dctrl = if (getRversion() > "3.4.4") c("keepNA", "niceNames") else "keepNA" + ) + on.exit(base::options(oop), add = TRUE) + if (run == FALSE) { + message(sprintf("\nStarting %s at the url shown below ...\nClick on the link or copy-and-paste it into\nyour browser's url bar to start", package)) + options(radiant.launch = "browser") + } else if (run == "browser" || run == "external") { + message(sprintf("\nStarting %s in the default browser", package)) + options(radiant.launch = "browser") + run <- TRUE + } else if (rstudioapi::getVersion() < "1.1") { + stop(sprintf("Rstudio version 1.1 or later required. Use %s::%s() to open %s in your default browser or download the latest version of Rstudio from https://posit.co/products/open-source/rstudio/", package, package, package)) + } else if (run == "viewer") { + message(sprintf("\nStarting %s in the Rstudio viewer ...\n\nUse %s::%s() to open %s in the default browser or %s::%s_window() in Rstudio to open %s in an Rstudio window", package, package, package, package, package, package, package)) + options(radiant.launch = "viewer") + run <- shiny::paneViewer(minHeight = "maximize") + } else if (run == "window") { + message(sprintf("\nStarting %s in an Rstudio window ...\n\nUse %s::%s() to open %s in the default browser or %s::%s_viewer() in Rstudio to open %s in the Rstudio viewer", package, package, package, package, package, package, package)) + os_type <- Sys.info()["sysname"] + if (os_type != "Darwin" && rstudioapi::getVersion() < "1.2") { + message(sprintf("\nUsing Radiant in an Rstudio Window works best in a newer version of Rstudio (i.e., version > 1.2). See https://dailies.rstudio.com/ for the latest version. Alternatively, use %s::%s_viewer()", package, package)) + } + options(radiant.launch = "window") + run <- get(".rs.invokeShinyWindowViewer") + } else { + message(sprintf("\nStarting %s in the default browser", package)) + options(radiant.launch = "browser") + run <- TRUE + } + + cat("\nRadiant is opensource and free to use. If you are a student or instructor using Radiant for a class, as a favor to the developer, please send an email to
 diff --git a/radiant.data/_pkgdown.yml b/radiant.data/_pkgdown.yml
new file mode 100644
index 0000000000000000000000000000000000000000..b43df3c6695aa7b762da63cb695bf350134123ad
--- /dev/null
+++ b/radiant.data/_pkgdown.yml
@@ -0,0 +1,265 @@
+url: https://radiant-rstats.github.io/radiant.data
+
+template:
+ params:
+ docsearch:
+ api_key: 311c7eff313b1f67999e5838086df74e
+ index_name: radiant_data
+
+navbar:
+ title: "radiant.data"
+ left:
+ - icon: fa-home fa-lg
+ href: index.html
+ - text: "Reference"
+ href: reference/index.html
+ - text: "Articles"
+ href: articles/index.html
+ - text: "Changelog"
+ href: news/index.html
+ - text: "Other Packages"
+ menu:
+ - text: "radiant"
+ href: https://radiant-rstats.github.io/radiant/
+ - text: "radiant.data"
+ href: https://radiant-rstats.github.io/radiant.data/
+ - text: "radiant.design"
+ href: https://radiant-rstats.github.io/radiant.design/
+ - text: "radiant.basics"
+ href: https://radiant-rstats.github.io/radiant.basics/
+ - text: "radiant.model"
+ href: https://radiant-rstats.github.io/radiant.model/
+ - text: "radiant.multivariate"
+ href: https://radiant-rstats.github.io/radiant.multivariate/
+ - text: "docker"
+ href: https://github.com/radiant-rstats/docker
+ right:
+ - icon: fa-twitter fa-lg
+ href: https://twitter.com/vrnijs
+ - icon: fa-github fa-lg
+ href: https://github.com/radiant-rstats
+
+reference:
+ - title: Data > Manage
+ desc: Functions used with Data > Manage
+ contents:
+ - choose_dir
+ - choose_files
+ - describe
+ - find_dropbox
+ - find_gdrive
+ - find_home
+ - find_project
+ - fix_names
+ - get_data
+ - load_clip
+ - parse_path
+ - read_files
+ - save_clip
+ - write_parquet
+ - to_fct
+ - title: Data > View
+ desc: Functions used with Data > View
+ contents:
+ - dtab
+ - dtab.data.frame
+ - filter_data
+ - make_arrange_cmd
+ - arrange_data
+ - slice_data
+ - search_data
+ - view_data
+ - title: Data > Visualize
+ desc: Function used with Data > Visualize
+ contents:
+ - visualize
+ - qscatter
+ - ggplotly
+ - subplot
+ - title: Data > Pivot
+ desc: Functions used with Data > Pivot
+ contents:
+ - pivotr
+ - summary.pivotr
+ - dtab.pivotr
+ - plot.pivotr
+ - title: Data > Explore
+ desc: Functions used with Data > Pivot
+ contents:
+ - explore
+ - summary.explore
+ - dtab.explore
+ - flip
+ - title: Data > Transform
+ desc: Functions used with Data > Transform
+ contents:
+ - as_character
+ - as_distance
+ - as_distance
+ - as_dmy
+ - as_dmy_hm
+ - as_dmy_hms
+ - as_duration
+ - as_factor
+ - as_hm
+ - as_hms
+ - as_integer
+ - as_mdy
+ - as_mdy_hm
+ - as_mdy_hms
+ - as_numeric
+ - as_ymd
+ - as_ymd_hm
+ - as_ymd_hms
+ - center
+ - cv
+ - inverse
+ - is.empty
+ - is_not
+ - is_double
+ - is_string
+ - level_list
+ - ln
+ - make_train
+ - month
+ - mutate_ext
+ - n_missing
+ - n_obs
+ - normalize
+ - make_vec
+ - me
+ - meprop
+ - modal
+ - p025
+ - p05
+ - p10
+ - p25
+ - p75
+ - p90
+ - p95
+ - p975
+ - prop
+ - refactor
+ - sdpop
+ - sdprop
+ - se
+ - seprop
+ - show_duplicated
+ - square
+ - standardize
+ - store
+ - table2data
+ - varpop
+ - varprop
+ - wday
+ - weighted.sd
+ - which.pmax
+ - which.pmin
+ - pfun
+ - psum
+ - pmean
+ - psd
+ - pvar
+ - pcv
+ - pp01
+ - pp025
+ - pp05
+ - pp25
+ - pp75
+ - pp95
+ - pp975
+ - pp99
+ - xtile
+ - title: Data > Combine
+ desc: Functions used with Data > Combine
+ contents:
+ - combine_data
+ - title: Report
+ desc: Functions used with Report > Rmd and Report > R
+ contents:
+ - fix_smart
+ - format_df
+ - format_nr
+ - round_df
+ - register
+ - deregister
+ - render
+ - render.datatables
+ - render.plotly
+ - title: Convenience functions
+ desc: Convenience functions
+ contents:
+ - add_class
+ - add_description
+ - get_class
+ - ci_label
+ - ci_perc
+ - copy_all
+ - copy_attr
+ - copy_from
+ - does_vary
+ - empty_level
+ - get_summary
+ - indexr
+ - install_webshot
+ - iterms
+ - qterms
+ - set_attr
+ - sig_stars
+ - sshh
+ - sshhr
+ - title: Starting radiant.data
+ desc: Functions used to start radiant shiny apps
+ contents:
+ - launch
+ - radiant.data
+ - radiant.data_url
+ - radiant.data_viewer
+ - radiant.data_window
+ - title: Re-exported
+ desc: Functions exported from other packages
+ contents:
+ - as_tibble
+ - tibble
+ - rownames_to_column
+ - glance
+ - tidy
+ - glue
+ - glue_collapse
+ - glue_data
+ - knit_print
+ - kurtosi
+ - skew
+ - title: Data sets
+ desc: Data sets bundled with radiant.data
+ contents:
+ - avengers
+ - diamonds
+ - publishers
+ - superheroes
+ - titanic
+ - title: Deprecated
+ desc: Deprecated
+ contents:
+ - radiant.data-deprecated
+ - store.pivotr
+ - store.explore
+articles:
+ - title: Data Menu
+ desc: >
+ These vignettes provide an introduction to the Data menu in radiant
+ contents:
+ - pkgdown/manage
+ - pkgdown/view
+ - pkgdown/visualize
+ - pkgdown/pivotr
+ - pkgdown/explore
+ - pkgdown/transform
+ - pkgdown/combine
+ - title: Report
+ contents:
+ - pkgdown/report_rmd
+ - pkgdown/report_r
+ - title: State
+ contents:
+ - pkgdown/state
diff --git a/radiant.data/build/build.R b/radiant.data/build/build.R
new file mode 100644
index 0000000000000000000000000000000000000000..9c0968d00465615a557f6acb1565bf9bcb256cd2
--- /dev/null
+++ b/radiant.data/build/build.R
@@ -0,0 +1,87 @@
+setwd(rstudioapi::getActiveProject())
+curr <- getwd()
+pkg <- basename(curr)
+
+## building package for mac and windows
+rv <- R.Version()
+rv <- paste(rv$major, substr(rv$minor, 1, 1), sep = ".")
+
+rvprompt <- readline(prompt = paste0("Running for R version: ", rv, ". Is that what you wanted y/n: "))
+if (grepl("[nN]", rvprompt)) stop("Change R-version")
+
+dirsrc <- "../minicran/src/contrib"
+
+if (rv < "3.4") {
+ dirmac <- fs::path("../minicran/bin/macosx/mavericks/contrib", rv)
+} else if (rv > "3.6") {
+ dirmac <- c(
+ fs::path("../minicran/bin/macosx/big-sur-arm64/contrib", rv),
+ fs::path("../minicran/bin/macosx/contrib", rv)
+ )
+} else {
+ dirmac <- fs::path("../minicran/bin/macosx/el-capitan/contrib", rv)
+}
+
+dirwin <- fs::path("../minicran/bin/windows/contrib", rv)
+
+if (!fs::file_exists(dirsrc)) fs::dir_create(dirsrc, recursive = TRUE)
+for (d in dirmac) {
+ if (!fs::file_exists(d)) fs::dir_create(d, recursive = TRUE)
+}
+if (!fs::file_exists(dirwin)) fs::dir_create(dirwin, recursive = TRUE)
+
+# delete older version of radiant
+rem_old <- function(pkg) {
+ unlink(paste0(dirsrc, "/", pkg, "*"))
+ for (d in dirmac) {
+ unlink(paste0(d, "/", pkg, "*"))
+ }
+ unlink(paste0(dirwin, "/", pkg, "*"))
+}
+
+sapply(pkg, rem_old)
+
+## avoid 'loaded namespace' stuff when building for mac
+system(paste0(Sys.which("R"), " -e \"setwd('", getwd(), "'); app <- '", pkg, "'; source('build/build_mac.R')\""))
+
+win <- readline(prompt = "Did you build on Windows? y/n: ")
+if (grepl("[yY]", win)) {
+
+ fl <- list.files(pattern = "*.zip", path = "~/Dropbox/r-packages", full.names = TRUE)
+ for (f in fl) {
+ file.copy(f, "~/gh/")
+ }
+ unlink(fl)
+
+ ## move packages to radiant_miniCRAN. must package in Windows first
+ # path <- normalizePath("../")
+ pth <- fs::path_abs("../")
+
+ sapply(list.files(pth, pattern = "*.tar.gz", full.names = TRUE), file.copy, dirsrc)
+ unlink("../*.tar.gz")
+ for (d in dirmac) {
+ sapply(list.files(pth, pattern = "*.tgz", full.names = TRUE), file.copy, d)
+ }
+ unlink("../*.tgz")
+ sapply(list.files(pth, pattern = "*.zip", full.names = TRUE), file.copy, dirwin)
+ unlink("../*.zip")
+
+ tools::write_PACKAGES(dirwin, type = "win.binary")
+ for (d in dirmac) {
+ tools::write_PACKAGES(d, type = "mac.binary")
+ }
+ tools::write_PACKAGES(dirsrc, type = "source")
+
+ # commit to repo
+ setwd("../minicran")
+ system("git add --all .")
+ mess <- paste0(pkg, " package update: ", format(Sys.Date(), format = "%m-%d-%Y"))
+ system(paste0("git commit -m '", mess, "'"))
+ system("git push")
+}
+
+setwd(curr)
+
+# remove.packages(c("radiant.model", "radiant.data"))
+# radiant.update::radiant.update()
+# install.packages("radiant.update")
diff --git a/radiant.data/build/build_mac.R b/radiant.data/build/build_mac.R
new file mode 100644
index 0000000000000000000000000000000000000000..1452bac080e154c24c6cd9acb6eef6c09a76c6ae
--- /dev/null
+++ b/radiant.data/build/build_mac.R
@@ -0,0 +1,6 @@
+## build for mac
+app <- basename(getwd())
+curr <- setwd("../")
+f <- devtools::build(app)
+system(paste0("R CMD INSTALL --build ", f))
+setwd(curr)
diff --git a/radiant.data/build/build_win.R b/radiant.data/build/build_win.R
new file mode 100644
index 0000000000000000000000000000000000000000..f988de18b876a6b5216dcf72656d7a8ad577144a
--- /dev/null
+++ b/radiant.data/build/build_win.R
@@ -0,0 +1,24 @@
+## build for windows
+rv <- R.Version()
+rv <- paste(rv$major, substr(rv$minor, 1, 1), sep = ".")
+
+rvprompt <- readline(prompt = paste0("Running for R version: ", rv, ". Is that what you wanted y/n: "))
+if (grepl("[nN]", rvprompt))
+ stop("Change R-version using Rstudio > Tools > Global Options > Rversion")
+
+## build for windows
+setwd(rstudioapi::getActiveProject())
+f <- devtools::build(binary = TRUE)
+devtools::install(upgrade = "never")
+
+f <- list.files(pattern = "*.zip", path = "../", full.names = TRUE)
+
+print(glue::glue("Copying: {f}"))
+file.copy(f, "C:/Users/vnijs/Dropbox/r-packages/", overwrite = TRUE)
+unlink(f)
+
+#options(repos = c(RSM = "https://radiant-rstats.github.io/minicran"))
+#install.packages("radiant.data", type = "binary")
+#remove.packages(c("radiant.data", "radiant.model"))
+#install.packages("radiant.update")
+#radiant.update::radiant.update()
diff --git a/radiant.data/cran-comments.md b/radiant.data/cran-comments.md
new file mode 100644
index 0000000000000000000000000000000000000000..1a3d0a8aedfe6721bf64858cea0e2db39f52b7c4
--- /dev/null
+++ b/radiant.data/cran-comments.md
@@ -0,0 +1,329 @@
+## Resubmission
+
+This is a resubmission. In this version I moved the arrow package to 'recommended' because of its size on macOS (>100MB)
+
+## Test environments
+
+* macOS, R 4.4.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+# Previous cran-comments
+
+## Resubmission
+
+This is a resubmission. In this version I require shiny version 1.8.1 or newer and have addressed a breaking change introduced in that version of shiny. See NEWS.md for details.
+
+## Test environments
+
+* macOS, R 4.4.0
+* macOS, R 4.3.2
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+
+## Resubmission
+
+This is a resubmission. In this version I require shiny version 1.8.0 which fixed a bug that caused issues in the radiant apps. See NEWS.md for details.
+
+## Test environments
+
+* macOS, R 4.3.2
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed a bug that caused problems for users on Windows with a space in their username. See NEWS.md for details.
+
+## Test environments
+
+* macOS, R 4.3.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this update I fixed a strange issue related the patchwork package. At least it was strange to me. In the code below `any` should not be needed. However, it seems that a patchwork object can have length == 1 and still have is.na return a vector of length > 1. Perhaps there are other libraries that have objects like this but I have never seen this before.
+
+My apologies for submitting a new version so soon after the previous version.
+
+```r
+length(x) == 0 || (length(x) == 1 && any(is.na(x)))
+```
+
+## Test environments
+
+* macOS, R 4.3.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this update I have added features and removed a bug. See NEWS.md.
+
+## Test environments
+
+* macOS, R 4.3.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+## Resubmission
+
+This is a resubmission. In this update I have added features and cleaned up code to avoid issues with markdown deprecation warnings. See NEWS.md.
+
+## Test environments
+
+* macOS, R 4.2.2
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+#
+
+## Resubmission
+
+This is a resubmission. In this update I have added features and cleaned up code to avoid issues with ggplot deprecation warnings. See NEWS.md. Also, URLs have been updated from Rstudio to Posit and the Radiant Documentation site is now back online and accessible.
+
+I have also tried to address the build issue connected to calibre. See note below.
+
+"This suggests you open a web browser in non interactive mode. Please use
+such calls only conditionally via
+
+if(interactive())
+
+Please fix and resubmit.
+
+Best,
+Uwe Ligges
+"
+
+## Test environments
+
+* macOS, R 4.2.2
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this update I have addressed three issues. See NEWS.md.
+
+## Test environments
+
+* macOS, R 4.2.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I fixed a dependency issue that is essential for correct functioning of the radiant.data shiny application. This feature is difficult to evaluate with automated testing and unfortunately I made a mistake in the submission earlier today. I uncovered the issue after upgrading to R 4.2.1. My apologies.
+
+## Test environments
+
+* macOS, R 4.2.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed a bug and added features (see NEWS.md for details).
+
+## Test environments
+
+* macOS, R 4.2.0
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed bugs and updated documentation (see NEWS.md for details). I also fixed an link issue in the documentation for sshh and sshhr
+
+## Test environments
+
+* macOS, R 4.2.0
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local Ubuntu 20.04, R 4.1.0
+* local Ubuntu 20.04 through WSL2, R 4.0.5
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+## Resubmission
+
+This is a resubmission. In this version I have address a problem linked to issue: https://github.com/yihui/knitr/issues/1864 There are also a number of changes that allow users to change the aesthetics of the app using `bslib` if available.
+
+## Test environments
+
+* local Ubuntu 20.04, R 4.1.0
+* local Ubuntu 20.04 through WSL2, R 4.0.5
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+# Previous cran-comments
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed issues related to updates in the `magrittr` and `readr` packages. I
+
+## Test environments
+
+* Ubuntu 20.04, R 4.0.3
+* win-builder (devel)
+* ubuntu "bionic" (on travis-ci), R release and devel
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have added back a feature that is now supported in dplyr 1.0.1 and made it easier to connect to Google Drive from the file-browser. I also, updated links that CRAN's automated checking listed.
+
+## Test environments
+
+* local OS X install, R 4.0.2
+* local Windows install, R 4.0.2
+* win-builder (devel)
+* ubuntu "bionic" (on travis-ci), R release and devel
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed a bug and removed a feature that no-longer works with dplyr 1.0.0 (see NEWS.md for details). Also, update the link to the ggplot2 documentation
+
+## Test environments
+
+* local OS X install, R 4.0.1
+* local Windows install, R 4.0.0
+* win-builder
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There is one NOTE about the number of imported non-default packages.
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local OS X install, R 3.6.3
+* local Windows install, R 3.6.2
+* ubuntu "trusty" (on travis-ci), R release and devel
+* win-builder
+
+## R CMD check results
+
+There were no ERRORs, WARNINGs, or NOTEs.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local OS X install, R 3.6.1
+* local Windows install, R 3.6.1
+* ubuntu "trusty" (on travis-ci), R release and devel
+* win-builder
+
+## R CMD check results
+
+There were no ERRORs, WARNINGs, or NOTEs.
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local OS X install, R 3.6.1
+* local Windows install, R 3.6.1
+* ubuntu "trusty" (on travis-ci), R release and devel
+* win-builder
+* rhub
+
+## R CMD check results
+
+There were no ERRORs, WARNINGs, or NOTEs.
+
+# Previous cran-comments
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local OS X install, R 3.5.2
+* local Windows install, R 3.5.2
+* ubuntu "trusty" (on travis-ci), R release and devel
+* win-builder
+
+## R CMD check results
+
+There were no ERRORs, WARNINGs, or NOTEs.
diff --git a/radiant.data/data/avengers.rda b/radiant.data/data/avengers.rda
new file mode 100644
index 0000000000000000000000000000000000000000..3356e7a0fb17abe99a953f22da37a8e8335d1ac6
Binary files /dev/null and b/radiant.data/data/avengers.rda differ
diff --git a/radiant.data/data/diamonds.rda b/radiant.data/data/diamonds.rda
new file mode 100644
index 0000000000000000000000000000000000000000..0a2e828a77d6904bdf2c7f2b82ae2d8290fe8049
Binary files /dev/null and b/radiant.data/data/diamonds.rda differ
diff --git a/radiant.data/data/publishers.rda b/radiant.data/data/publishers.rda
new file mode 100644
index 0000000000000000000000000000000000000000..764affc023f365f67e89c87664ecb6234b77c1b3
Binary files /dev/null and b/radiant.data/data/publishers.rda differ
diff --git a/radiant.data/data/superheroes.rda b/radiant.data/data/superheroes.rda
new file mode 100644
index 0000000000000000000000000000000000000000..edcdc27ca2c4fba5b8fe2cf124101419f7726386
Binary files /dev/null and b/radiant.data/data/superheroes.rda differ
diff --git a/radiant.data/data/titanic.rda b/radiant.data/data/titanic.rda
new file mode 100644
index 0000000000000000000000000000000000000000..ecb4584b532e187340552fa3e5df3bbae6407f48
Binary files /dev/null and b/radiant.data/data/titanic.rda differ
diff --git a/radiant.data/docs/404.html b/radiant.data/docs/404.html
new file mode 100644
index 0000000000000000000000000000000000000000..c64a8f36fe02fc1214dd80a8f4f241cedd5cc179
--- /dev/null
+++ b/radiant.data/docs/404.html
@@ -0,0 +1,167 @@
+
+
+
+
+
+
+
+
diff --git a/radiant.data/_pkgdown.yml b/radiant.data/_pkgdown.yml
new file mode 100644
index 0000000000000000000000000000000000000000..b43df3c6695aa7b762da63cb695bf350134123ad
--- /dev/null
+++ b/radiant.data/_pkgdown.yml
@@ -0,0 +1,265 @@
+url: https://radiant-rstats.github.io/radiant.data
+
+template:
+ params:
+ docsearch:
+ api_key: 311c7eff313b1f67999e5838086df74e
+ index_name: radiant_data
+
+navbar:
+ title: "radiant.data"
+ left:
+ - icon: fa-home fa-lg
+ href: index.html
+ - text: "Reference"
+ href: reference/index.html
+ - text: "Articles"
+ href: articles/index.html
+ - text: "Changelog"
+ href: news/index.html
+ - text: "Other Packages"
+ menu:
+ - text: "radiant"
+ href: https://radiant-rstats.github.io/radiant/
+ - text: "radiant.data"
+ href: https://radiant-rstats.github.io/radiant.data/
+ - text: "radiant.design"
+ href: https://radiant-rstats.github.io/radiant.design/
+ - text: "radiant.basics"
+ href: https://radiant-rstats.github.io/radiant.basics/
+ - text: "radiant.model"
+ href: https://radiant-rstats.github.io/radiant.model/
+ - text: "radiant.multivariate"
+ href: https://radiant-rstats.github.io/radiant.multivariate/
+ - text: "docker"
+ href: https://github.com/radiant-rstats/docker
+ right:
+ - icon: fa-twitter fa-lg
+ href: https://twitter.com/vrnijs
+ - icon: fa-github fa-lg
+ href: https://github.com/radiant-rstats
+
+reference:
+ - title: Data > Manage
+ desc: Functions used with Data > Manage
+ contents:
+ - choose_dir
+ - choose_files
+ - describe
+ - find_dropbox
+ - find_gdrive
+ - find_home
+ - find_project
+ - fix_names
+ - get_data
+ - load_clip
+ - parse_path
+ - read_files
+ - save_clip
+ - write_parquet
+ - to_fct
+ - title: Data > View
+ desc: Functions used with Data > View
+ contents:
+ - dtab
+ - dtab.data.frame
+ - filter_data
+ - make_arrange_cmd
+ - arrange_data
+ - slice_data
+ - search_data
+ - view_data
+ - title: Data > Visualize
+ desc: Function used with Data > Visualize
+ contents:
+ - visualize
+ - qscatter
+ - ggplotly
+ - subplot
+ - title: Data > Pivot
+ desc: Functions used with Data > Pivot
+ contents:
+ - pivotr
+ - summary.pivotr
+ - dtab.pivotr
+ - plot.pivotr
+ - title: Data > Explore
+ desc: Functions used with Data > Pivot
+ contents:
+ - explore
+ - summary.explore
+ - dtab.explore
+ - flip
+ - title: Data > Transform
+ desc: Functions used with Data > Transform
+ contents:
+ - as_character
+ - as_distance
+ - as_distance
+ - as_dmy
+ - as_dmy_hm
+ - as_dmy_hms
+ - as_duration
+ - as_factor
+ - as_hm
+ - as_hms
+ - as_integer
+ - as_mdy
+ - as_mdy_hm
+ - as_mdy_hms
+ - as_numeric
+ - as_ymd
+ - as_ymd_hm
+ - as_ymd_hms
+ - center
+ - cv
+ - inverse
+ - is.empty
+ - is_not
+ - is_double
+ - is_string
+ - level_list
+ - ln
+ - make_train
+ - month
+ - mutate_ext
+ - n_missing
+ - n_obs
+ - normalize
+ - make_vec
+ - me
+ - meprop
+ - modal
+ - p025
+ - p05
+ - p10
+ - p25
+ - p75
+ - p90
+ - p95
+ - p975
+ - prop
+ - refactor
+ - sdpop
+ - sdprop
+ - se
+ - seprop
+ - show_duplicated
+ - square
+ - standardize
+ - store
+ - table2data
+ - varpop
+ - varprop
+ - wday
+ - weighted.sd
+ - which.pmax
+ - which.pmin
+ - pfun
+ - psum
+ - pmean
+ - psd
+ - pvar
+ - pcv
+ - pp01
+ - pp025
+ - pp05
+ - pp25
+ - pp75
+ - pp95
+ - pp975
+ - pp99
+ - xtile
+ - title: Data > Combine
+ desc: Functions used with Data > Combine
+ contents:
+ - combine_data
+ - title: Report
+ desc: Functions used with Report > Rmd and Report > R
+ contents:
+ - fix_smart
+ - format_df
+ - format_nr
+ - round_df
+ - register
+ - deregister
+ - render
+ - render.datatables
+ - render.plotly
+ - title: Convenience functions
+ desc: Convenience functions
+ contents:
+ - add_class
+ - add_description
+ - get_class
+ - ci_label
+ - ci_perc
+ - copy_all
+ - copy_attr
+ - copy_from
+ - does_vary
+ - empty_level
+ - get_summary
+ - indexr
+ - install_webshot
+ - iterms
+ - qterms
+ - set_attr
+ - sig_stars
+ - sshh
+ - sshhr
+ - title: Starting radiant.data
+ desc: Functions used to start radiant shiny apps
+ contents:
+ - launch
+ - radiant.data
+ - radiant.data_url
+ - radiant.data_viewer
+ - radiant.data_window
+ - title: Re-exported
+ desc: Functions exported from other packages
+ contents:
+ - as_tibble
+ - tibble
+ - rownames_to_column
+ - glance
+ - tidy
+ - glue
+ - glue_collapse
+ - glue_data
+ - knit_print
+ - kurtosi
+ - skew
+ - title: Data sets
+ desc: Data sets bundled with radiant.data
+ contents:
+ - avengers
+ - diamonds
+ - publishers
+ - superheroes
+ - titanic
+ - title: Deprecated
+ desc: Deprecated
+ contents:
+ - radiant.data-deprecated
+ - store.pivotr
+ - store.explore
+articles:
+ - title: Data Menu
+ desc: >
+ These vignettes provide an introduction to the Data menu in radiant
+ contents:
+ - pkgdown/manage
+ - pkgdown/view
+ - pkgdown/visualize
+ - pkgdown/pivotr
+ - pkgdown/explore
+ - pkgdown/transform
+ - pkgdown/combine
+ - title: Report
+ contents:
+ - pkgdown/report_rmd
+ - pkgdown/report_r
+ - title: State
+ contents:
+ - pkgdown/state
diff --git a/radiant.data/build/build.R b/radiant.data/build/build.R
new file mode 100644
index 0000000000000000000000000000000000000000..9c0968d00465615a557f6acb1565bf9bcb256cd2
--- /dev/null
+++ b/radiant.data/build/build.R
@@ -0,0 +1,87 @@
+setwd(rstudioapi::getActiveProject())
+curr <- getwd()
+pkg <- basename(curr)
+
+## building package for mac and windows
+rv <- R.Version()
+rv <- paste(rv$major, substr(rv$minor, 1, 1), sep = ".")
+
+rvprompt <- readline(prompt = paste0("Running for R version: ", rv, ". Is that what you wanted y/n: "))
+if (grepl("[nN]", rvprompt)) stop("Change R-version")
+
+dirsrc <- "../minicran/src/contrib"
+
+if (rv < "3.4") {
+ dirmac <- fs::path("../minicran/bin/macosx/mavericks/contrib", rv)
+} else if (rv > "3.6") {
+ dirmac <- c(
+ fs::path("../minicran/bin/macosx/big-sur-arm64/contrib", rv),
+ fs::path("../minicran/bin/macosx/contrib", rv)
+ )
+} else {
+ dirmac <- fs::path("../minicran/bin/macosx/el-capitan/contrib", rv)
+}
+
+dirwin <- fs::path("../minicran/bin/windows/contrib", rv)
+
+if (!fs::file_exists(dirsrc)) fs::dir_create(dirsrc, recursive = TRUE)
+for (d in dirmac) {
+ if (!fs::file_exists(d)) fs::dir_create(d, recursive = TRUE)
+}
+if (!fs::file_exists(dirwin)) fs::dir_create(dirwin, recursive = TRUE)
+
+# delete older version of radiant
+rem_old <- function(pkg) {
+ unlink(paste0(dirsrc, "/", pkg, "*"))
+ for (d in dirmac) {
+ unlink(paste0(d, "/", pkg, "*"))
+ }
+ unlink(paste0(dirwin, "/", pkg, "*"))
+}
+
+sapply(pkg, rem_old)
+
+## avoid 'loaded namespace' stuff when building for mac
+system(paste0(Sys.which("R"), " -e \"setwd('", getwd(), "'); app <- '", pkg, "'; source('build/build_mac.R')\""))
+
+win <- readline(prompt = "Did you build on Windows? y/n: ")
+if (grepl("[yY]", win)) {
+
+ fl <- list.files(pattern = "*.zip", path = "~/Dropbox/r-packages", full.names = TRUE)
+ for (f in fl) {
+ file.copy(f, "~/gh/")
+ }
+ unlink(fl)
+
+ ## move packages to radiant_miniCRAN. must package in Windows first
+ # path <- normalizePath("../")
+ pth <- fs::path_abs("../")
+
+ sapply(list.files(pth, pattern = "*.tar.gz", full.names = TRUE), file.copy, dirsrc)
+ unlink("../*.tar.gz")
+ for (d in dirmac) {
+ sapply(list.files(pth, pattern = "*.tgz", full.names = TRUE), file.copy, d)
+ }
+ unlink("../*.tgz")
+ sapply(list.files(pth, pattern = "*.zip", full.names = TRUE), file.copy, dirwin)
+ unlink("../*.zip")
+
+ tools::write_PACKAGES(dirwin, type = "win.binary")
+ for (d in dirmac) {
+ tools::write_PACKAGES(d, type = "mac.binary")
+ }
+ tools::write_PACKAGES(dirsrc, type = "source")
+
+ # commit to repo
+ setwd("../minicran")
+ system("git add --all .")
+ mess <- paste0(pkg, " package update: ", format(Sys.Date(), format = "%m-%d-%Y"))
+ system(paste0("git commit -m '", mess, "'"))
+ system("git push")
+}
+
+setwd(curr)
+
+# remove.packages(c("radiant.model", "radiant.data"))
+# radiant.update::radiant.update()
+# install.packages("radiant.update")
diff --git a/radiant.data/build/build_mac.R b/radiant.data/build/build_mac.R
new file mode 100644
index 0000000000000000000000000000000000000000..1452bac080e154c24c6cd9acb6eef6c09a76c6ae
--- /dev/null
+++ b/radiant.data/build/build_mac.R
@@ -0,0 +1,6 @@
+## build for mac
+app <- basename(getwd())
+curr <- setwd("../")
+f <- devtools::build(app)
+system(paste0("R CMD INSTALL --build ", f))
+setwd(curr)
diff --git a/radiant.data/build/build_win.R b/radiant.data/build/build_win.R
new file mode 100644
index 0000000000000000000000000000000000000000..f988de18b876a6b5216dcf72656d7a8ad577144a
--- /dev/null
+++ b/radiant.data/build/build_win.R
@@ -0,0 +1,24 @@
+## build for windows
+rv <- R.Version()
+rv <- paste(rv$major, substr(rv$minor, 1, 1), sep = ".")
+
+rvprompt <- readline(prompt = paste0("Running for R version: ", rv, ". Is that what you wanted y/n: "))
+if (grepl("[nN]", rvprompt))
+ stop("Change R-version using Rstudio > Tools > Global Options > Rversion")
+
+## build for windows
+setwd(rstudioapi::getActiveProject())
+f <- devtools::build(binary = TRUE)
+devtools::install(upgrade = "never")
+
+f <- list.files(pattern = "*.zip", path = "../", full.names = TRUE)
+
+print(glue::glue("Copying: {f}"))
+file.copy(f, "C:/Users/vnijs/Dropbox/r-packages/", overwrite = TRUE)
+unlink(f)
+
+#options(repos = c(RSM = "https://radiant-rstats.github.io/minicran"))
+#install.packages("radiant.data", type = "binary")
+#remove.packages(c("radiant.data", "radiant.model"))
+#install.packages("radiant.update")
+#radiant.update::radiant.update()
diff --git a/radiant.data/cran-comments.md b/radiant.data/cran-comments.md
new file mode 100644
index 0000000000000000000000000000000000000000..1a3d0a8aedfe6721bf64858cea0e2db39f52b7c4
--- /dev/null
+++ b/radiant.data/cran-comments.md
@@ -0,0 +1,329 @@
+## Resubmission
+
+This is a resubmission. In this version I moved the arrow package to 'recommended' because of its size on macOS (>100MB)
+
+## Test environments
+
+* macOS, R 4.4.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+# Previous cran-comments
+
+## Resubmission
+
+This is a resubmission. In this version I require shiny version 1.8.1 or newer and have addressed a breaking change introduced in that version of shiny. See NEWS.md for details.
+
+## Test environments
+
+* macOS, R 4.4.0
+* macOS, R 4.3.2
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+
+## Resubmission
+
+This is a resubmission. In this version I require shiny version 1.8.0 which fixed a bug that caused issues in the radiant apps. See NEWS.md for details.
+
+## Test environments
+
+* macOS, R 4.3.2
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed a bug that caused problems for users on Windows with a space in their username. See NEWS.md for details.
+
+## Test environments
+
+* macOS, R 4.3.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this update I fixed a strange issue related the patchwork package. At least it was strange to me. In the code below `any` should not be needed. However, it seems that a patchwork object can have length == 1 and still have is.na return a vector of length > 1. Perhaps there are other libraries that have objects like this but I have never seen this before.
+
+My apologies for submitting a new version so soon after the previous version.
+
+```r
+length(x) == 0 || (length(x) == 1 && any(is.na(x)))
+```
+
+## Test environments
+
+* macOS, R 4.3.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this update I have added features and removed a bug. See NEWS.md.
+
+## Test environments
+
+* macOS, R 4.3.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+## Resubmission
+
+This is a resubmission. In this update I have added features and cleaned up code to avoid issues with markdown deprecation warnings. See NEWS.md.
+
+## Test environments
+
+* macOS, R 4.2.2
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+#
+
+## Resubmission
+
+This is a resubmission. In this update I have added features and cleaned up code to avoid issues with ggplot deprecation warnings. See NEWS.md. Also, URLs have been updated from Rstudio to Posit and the Radiant Documentation site is now back online and accessible.
+
+I have also tried to address the build issue connected to calibre. See note below.
+
+"This suggests you open a web browser in non interactive mode. Please use
+such calls only conditionally via
+
+if(interactive())
+
+Please fix and resubmit.
+
+Best,
+Uwe Ligges
+"
+
+## Test environments
+
+* macOS, R 4.2.2
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this update I have addressed three issues. See NEWS.md.
+
+## Test environments
+
+* macOS, R 4.2.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I fixed a dependency issue that is essential for correct functioning of the radiant.data shiny application. This feature is difficult to evaluate with automated testing and unfortunately I made a mistake in the submission earlier today. I uncovered the issue after upgrading to R 4.2.1. My apologies.
+
+## Test environments
+
+* macOS, R 4.2.1
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed a bug and added features (see NEWS.md for details).
+
+## Test environments
+
+* macOS, R 4.2.0
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed bugs and updated documentation (see NEWS.md for details). I also fixed an link issue in the documentation for sshh and sshhr
+
+## Test environments
+
+* macOS, R 4.2.0
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local Ubuntu 20.04, R 4.1.0
+* local Ubuntu 20.04 through WSL2, R 4.0.5
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+## Resubmission
+
+This is a resubmission. In this version I have address a problem linked to issue: https://github.com/yihui/knitr/issues/1864 There are also a number of changes that allow users to change the aesthetics of the app using `bslib` if available.
+
+## Test environments
+
+* local Ubuntu 20.04, R 4.1.0
+* local Ubuntu 20.04 through WSL2, R 4.0.5
+* win-builder (devel)
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+# Previous cran-comments
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed issues related to updates in the `magrittr` and `readr` packages. I
+
+## Test environments
+
+* Ubuntu 20.04, R 4.0.3
+* win-builder (devel)
+* ubuntu "bionic" (on travis-ci), R release and devel
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have added back a feature that is now supported in dplyr 1.0.1 and made it easier to connect to Google Drive from the file-browser. I also, updated links that CRAN's automated checking listed.
+
+## Test environments
+
+* local OS X install, R 4.0.2
+* local Windows install, R 4.0.2
+* win-builder (devel)
+* ubuntu "bionic" (on travis-ci), R release and devel
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There was one NOTE related to the number of non-standard dependencies. However, this note is not easily addressed without substantially inconveniencing users that rely on the web (shiny) interface available for radiant.data.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed a bug and removed a feature that no-longer works with dplyr 1.0.0 (see NEWS.md for details). Also, update the link to the ggplot2 documentation
+
+## Test environments
+
+* local OS X install, R 4.0.1
+* local Windows install, R 4.0.0
+* win-builder
+
+## R CMD check results
+
+There were no ERRORs or WARNINGs. There is one NOTE about the number of imported non-default packages.
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local OS X install, R 3.6.3
+* local Windows install, R 3.6.2
+* ubuntu "trusty" (on travis-ci), R release and devel
+* win-builder
+
+## R CMD check results
+
+There were no ERRORs, WARNINGs, or NOTEs.
+
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local OS X install, R 3.6.1
+* local Windows install, R 3.6.1
+* ubuntu "trusty" (on travis-ci), R release and devel
+* win-builder
+
+## R CMD check results
+
+There were no ERRORs, WARNINGs, or NOTEs.
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local OS X install, R 3.6.1
+* local Windows install, R 3.6.1
+* ubuntu "trusty" (on travis-ci), R release and devel
+* win-builder
+* rhub
+
+## R CMD check results
+
+There were no ERRORs, WARNINGs, or NOTEs.
+
+# Previous cran-comments
+
+## Resubmission
+
+This is a resubmission. In this version I have fixed several bugs and added several new features (see NEWS.md for details).
+
+## Test environments
+
+* local OS X install, R 3.5.2
+* local Windows install, R 3.5.2
+* ubuntu "trusty" (on travis-ci), R release and devel
+* win-builder
+
+## R CMD check results
+
+There were no ERRORs, WARNINGs, or NOTEs.
diff --git a/radiant.data/data/avengers.rda b/radiant.data/data/avengers.rda
new file mode 100644
index 0000000000000000000000000000000000000000..3356e7a0fb17abe99a953f22da37a8e8335d1ac6
Binary files /dev/null and b/radiant.data/data/avengers.rda differ
diff --git a/radiant.data/data/diamonds.rda b/radiant.data/data/diamonds.rda
new file mode 100644
index 0000000000000000000000000000000000000000..0a2e828a77d6904bdf2c7f2b82ae2d8290fe8049
Binary files /dev/null and b/radiant.data/data/diamonds.rda differ
diff --git a/radiant.data/data/publishers.rda b/radiant.data/data/publishers.rda
new file mode 100644
index 0000000000000000000000000000000000000000..764affc023f365f67e89c87664ecb6234b77c1b3
Binary files /dev/null and b/radiant.data/data/publishers.rda differ
diff --git a/radiant.data/data/superheroes.rda b/radiant.data/data/superheroes.rda
new file mode 100644
index 0000000000000000000000000000000000000000..edcdc27ca2c4fba5b8fe2cf124101419f7726386
Binary files /dev/null and b/radiant.data/data/superheroes.rda differ
diff --git a/radiant.data/data/titanic.rda b/radiant.data/data/titanic.rda
new file mode 100644
index 0000000000000000000000000000000000000000..ecb4584b532e187340552fa3e5df3bbae6407f48
Binary files /dev/null and b/radiant.data/data/titanic.rda differ
diff --git a/radiant.data/docs/404.html b/radiant.data/docs/404.html
new file mode 100644
index 0000000000000000000000000000000000000000..c64a8f36fe02fc1214dd80a8f4f241cedd5cc179
--- /dev/null
+++ b/radiant.data/docs/404.html
@@ -0,0 +1,167 @@
+
+
+
+
+
+
+
+Page not found (404)
+License
+radiant.data is licensed under AGPL3 (see https://tldrlegal.com/license/gnu-affero-general-public-license-v3-(agpl-3.0) and https://www.r-project.org/Licenses/AGPL-3). The radiant.data help files and images are licensed under the creative commons attribution and share-alike license CC-BY-SA (https://creativecommons.org/licenses/by-sa/4.0/legalcode).
+
+As a summary, the AGPLv3 license requires, attribution, including copyright and license information in copies of the software, stating changes if the code is modified, and disclosure of all source code. Details are in the COPYING file.
+
+If you are interested in using radiant.data or other radiant packages please email me at radiant@rady.ucsd.edu
+
+====================================================================
+
+Creative Commons Attribution-ShareAlike 4.0 International Public License
+By exercising the Licensed Rights (defined below), You accept and agree to be bound by the terms and conditions of this Creative Commons Attribution-ShareAlike 4.0 International Public License ("Public License"). To the extent this Public License may be interpreted as a contract, You are granted the Licensed Rights in consideration of Your acceptance of these terms and conditions, and the Licensor grants You such rights in consideration of benefits the Licensor receives from making the Licensed Material available under these terms and conditions.
+
+Section 1 – Definitions.
+
+Adapted Material means material subject to Copyright and Similar Rights that is derived from or based upon the Licensed Material and in which the Licensed Material is translated, altered, arranged, transformed, or otherwise modified in a manner requiring permission under the Copyright and Similar Rights held by the Licensor. For purposes of this Public License, where the Licensed Material is a musical work, performance, or sound recording, Adapted Material is always produced where the Licensed Material is synched in timed relation with a moving image.
+Adapter's License means the license You apply to Your Copyright and Similar Rights in Your contributions to Adapted Material in accordance with the terms and conditions of this Public License.
+BY-SA Compatible License means a license listed at creativecommons.org/compatiblelicenses, approved by Creative Commons as essentially the equivalent of this Public License.
+Copyright and Similar Rights means copyright and/or similar rights closely related to copyright including, without limitation, performance, broadcast, sound recording, and Sui Generis Database Rights, without regard to how the rights are labeled or categorized. For purposes of this Public License, the rights specified in Section 2(b)(1)-(2) are not Copyright and Similar Rights.
+Effective Technological Measures means those measures that, in the absence of proper authority, may not be circumvented under laws fulfilling obligations under Article 11 of the WIPO Copyright Treaty adopted on December 20, 1996, and/or similar international agreements.
+Exceptions and Limitations means fair use, fair dealing, and/or any other exception or limitation to Copyright and Similar Rights that applies to Your use of the Licensed Material.
+License Elements means the license attributes listed in the name of a Creative Commons Public License. The License Elements of this Public License are Attribution and ShareAlike.
+Licensed Material means the artistic or literary work, database, or other material to which the Licensor applied this Public License.
+Licensed Rights means the rights granted to You subject to the terms and conditions of this Public License, which are limited to all Copyright and Similar Rights that apply to Your use of the Licensed Material and that the Licensor has authority to license.
+Licensor means the individual(s) or entity(ies) granting rights under this Public License.
+Share means to provide material to the public by any means or process that requires permission under the Licensed Rights, such as reproduction, public display, public performance, distribution, dissemination, communication, or importation, and to make material available to the public including in ways that members of the public may access the material from a place and at a time individually chosen by them.
+Sui Generis Database Rights means rights other than copyright resulting from Directive 96/9/EC of the European Parliament and of the Council of 11 March 1996 on the legal protection of databases, as amended and/or succeeded, as well as other essentially equivalent rights anywhere in the world.
+You means the individual or entity exercising the Licensed Rights under this Public License. Your has a corresponding meaning.
+Section 2 – Scope.
+
+License grant.
+Subject to the terms and conditions of this Public License, the Licensor hereby grants You a worldwide, royalty-free, non-sublicensable, non-exclusive, irrevocable license to exercise the Licensed Rights in the Licensed Material to:
+reproduce and Share the Licensed Material, in whole or in part; and
+produce, reproduce, and Share Adapted Material.
+Exceptions and Limitations. For the avoidance of doubt, where Exceptions and Limitations apply to Your use, this Public License does not apply, and You do not need to comply with its terms and conditions.
+Term. The term of this Public License is specified in Section 6(a).
+Media and formats; technical modifications allowed. The Licensor authorizes You to exercise the Licensed Rights in all media and formats whether now known or hereafter created, and to make technical modifications necessary to do so. The Licensor waives and/or agrees not to assert any right or authority to forbid You from making technical modifications necessary to exercise the Licensed Rights, including technical modifications necessary to circumvent Effective Technological Measures. For purposes of this Public License, simply making modifications authorized by this Section 2(a)(4) never produces Adapted Material.
+Downstream recipients.
+Offer from the Licensor – Licensed Material. Every recipient of the Licensed Material automatically receives an offer from the Licensor to exercise the Licensed Rights under the terms and conditions of this Public License.
+Additional offer from the Licensor – Adapted Material. Every recipient of Adapted Material from You automatically receives an offer from the Licensor to exercise the Licensed Rights in the Adapted Material under the conditions of the Adapter’s License You apply.
+No downstream restrictions. You may not offer or impose any additional or different terms or conditions on, or apply any Effective Technological Measures to, the Licensed Material if doing so restricts exercise of the Licensed Rights by any recipient of the Licensed Material.
+No endorsement. Nothing in this Public License constitutes or may be construed as permission to assert or imply that You are, or that Your use of the Licensed Material is, connected with, or sponsored, endorsed, or granted official status by, the Licensor or others designated to receive attribution as provided in Section 3(a)(1)(A)(i).
+Other rights.
+
+Moral rights, such as the right of integrity, are not licensed under this Public License, nor are publicity, privacy, and/or other similar personality rights; however, to the extent possible, the Licensor waives and/or agrees not to assert any such rights held by the Licensor to the limited extent necessary to allow You to exercise the Licensed Rights, but not otherwise.
+Patent and trademark rights are not licensed under this Public License.
+To the extent possible, the Licensor waives any right to collect royalties from You for the exercise of the Licensed Rights, whether directly or through a collecting society under any voluntary or waivable statutory or compulsory licensing scheme. In all other cases the Licensor expressly reserves any right to collect such royalties.
+Section 3 – License Conditions.
+
+Your exercise of the Licensed Rights is expressly made subject to the following conditions.
+
+Attribution.
+
+If You Share the Licensed Material (including in modified form), You must:
+
+retain the following if it is supplied by the Licensor with the Licensed Material:
+identification of the creator(s) of the Licensed Material and any others designated to receive attribution, in any reasonable manner requested by the Licensor (including by pseudonym if designated);
+a copyright notice;
+a notice that refers to this Public License;
+a notice that refers to the disclaimer of warranties;
+a URI or hyperlink to the Licensed Material to the extent reasonably practicable;
+indicate if You modified the Licensed Material and retain an indication of any previous modifications; and
+indicate the Licensed Material is licensed under this Public License, and include the text of, or the URI or hyperlink to, this Public License.
+You may satisfy the conditions in Section 3(a)(1) in any reasonable manner based on the medium, means, and context in which You Share the Licensed Material. For example, it may be reasonable to satisfy the conditions by providing a URI or hyperlink to a resource that includes the required information.
+If requested by the Licensor, You must remove any of the information required by Section 3(a)(1)(A) to the extent reasonably practicable.
+ShareAlike.
+In addition to the conditions in Section 3(a), if You Share Adapted Material You produce, the following conditions also apply.
+
+The Adapter’s License You apply must be a Creative Commons license with the same License Elements, this version or later, or a BY-SA Compatible License.
+You must include the text of, or the URI or hyperlink to, the Adapter's License You apply. You may satisfy this condition in any reasonable manner based on the medium, means, and context in which You Share Adapted Material.
+You may not offer or impose any additional or different terms or conditions on, or apply any Effective Technological Measures to, Adapted Material that restrict exercise of the rights granted under the Adapter's License You apply.
+Section 4 – Sui Generis Database Rights.
+
+Where the Licensed Rights include Sui Generis Database Rights that apply to Your use of the Licensed Material:
+
+for the avoidance of doubt, Section 2(a)(1) grants You the right to extract, reuse, reproduce, and Share all or a substantial portion of the contents of the database;
+if You include all or a substantial portion of the database contents in a database in which You have Sui Generis Database Rights, then the database in which You have Sui Generis Database Rights (but not its individual contents) is Adapted Material, including for purposes of Section 3(b); and
+You must comply with the conditions in Section 3(a) if You Share all or a substantial portion of the contents of the database.
+For the avoidance of doubt, this Section 4 supplements and does not replace Your obligations under this Public License where the Licensed Rights include other Copyright and Similar Rights.
+Section 5 – Disclaimer of Warranties and Limitation of Liability.
+
+Unless otherwise separately undertaken by the Licensor, to the extent possible, the Licensor offers the Licensed Material as-is and as-available, and makes no representations or warranties of any kind concerning the Licensed Material, whether express, implied, statutory, or other. This includes, without limitation, warranties of title, merchantability, fitness for a particular purpose, non-infringement, absence of latent or other defects, accuracy, or the presence or absence of errors, whether or not known or discoverable. Where disclaimers of warranties are not allowed in full or in part, this disclaimer may not apply to You.
+To the extent possible, in no event will the Licensor be liable to You on any legal theory (including, without limitation, negligence) or otherwise for any direct, special, indirect, incidental, consequential, punitive, exemplary, or other losses, costs, expenses, or damages arising out of this Public License or use of the Licensed Material, even if the Licensor has been advised of the possibility of such losses, costs, expenses, or damages. Where a limitation of liability is not allowed in full or in part, this limitation may not apply to You.
+The disclaimer of warranties and limitation of liability provided above shall be interpreted in a manner that, to the extent possible, most closely approximates an absolute disclaimer and waiver of all liability.
+Section 6 – Term and Termination.
+
+This Public License applies for the term of the Copyright and Similar Rights licensed here. However, if You fail to comply with this Public License, then Your rights under this Public License terminate automatically.
+Where Your right to use the Licensed Material has terminated under Section 6(a), it reinstates:
+
+automatically as of the date the violation is cured, provided it is cured within 30 days of Your discovery of the violation; or
+upon express reinstatement by the Licensor.
+For the avoidance of doubt, this Section 6(b) does not affect any right the Licensor may have to seek remedies for Your violations of this Public License.
+For the avoidance of doubt, the Licensor may also offer the Licensed Material under separate terms or conditions or stop distributing the Licensed Material at any time; however, doing so will not terminate this Public License.
+Sections 1, 5, 6, 7, and 8 survive termination of this Public License.
+Section 7 – Other Terms and Conditions.
+
+The Licensor shall not be bound by any additional or different terms or conditions communicated by You unless expressly agreed.
+Any arrangements, understandings, or agreements regarding the Licensed Material not stated herein are separate from and independent of the terms and conditions of this Public License.
+Section 8 – Interpretation.
+
+For the avoidance of doubt, this Public License does not, and shall not be interpreted to, reduce, limit, restrict, or impose conditions on any use of the Licensed Material that could lawfully be made without permission under this Public License.
+To the extent possible, if any provision of this Public License is deemed unenforceable, it shall be automatically reformed to the minimum extent necessary to make it enforceable. If the provision cannot be reformed, it shall be severed from this Public License without affecting the enforceability of the remaining terms and conditions.
+No term or condition of this Public License will be waived and no failure to comply consented to unless expressly agreed to by the Licensor.
+Nothing in this Public License constitutes or may be interpreted as a limitation upon, or waiver of, any privileges and immunities that apply to the Licensor or You, including from the legal processes of any jurisdiction or authority.
+
+
+ Articles
+Data Menu
+These vignettes provide an introduction to the Data menu in radiant
+ +- Loading and Saving data (Data > Manage) +
- +
- View data in an interactive table (Data > View) +
- +
- Visualize data (Data > Visualize) +
- +
- Create pivot tables (Data > Pivot) +
- +
- Summarize and explore your data (Data > Explore) +
- +
- Transform variables (Data > Transform) +
- +
- Combine data sets (Data > Combine) +
- +
Report
+ + +State
+ + +Combine data sets (Data > Combine)
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/combine.Rmd
+ combine.Rmd++Combine two datasets
+
There are six join (or merge) options available in +Radiant from the +dplyr +package developed by Hadley Wickham et.al.
+The examples below are adapted from the
+Cheatsheet
+for dplyr join functions by
+Jenny Bryan
+and focus on three small datasets, superheroes,
+publishers, and avengers, to illustrate the
+different join types and other ways to combine datasets in R
+and Radiant. The data are also available in csv format through the links
+below:
| +name + | ++alignment + | ++gender + | ++publisher + | +
|---|---|---|---|
| +Magneto + | ++bad + | ++male + | ++Marvel + | +
| +Storm + | ++good + | ++female + | ++Marvel + | +
| +Mystique + | ++bad + | ++female + | ++Marvel + | +
| +Batman + | ++good + | ++male + | ++DC + | +
| +Joker + | ++bad + | ++male + | ++DC + | +
| +Catwoman + | ++bad + | ++female + | ++DC + | +
| +Hellboy + | ++good + | ++male + | ++Dark Horse Comics + | +
| +publisher + | ++yr_founded + | +
|---|---|
| +DC + | ++1934 + | +
| +Marvel + | ++1939 + | +
| +Image + | ++1992 + | +
In the screen-shot of the Data > Combine tab below we see
+the two datasets. The tables share the variable publisher which
+is automatically selected for the join. Different join options are
+available from the Combine type dropdown. You can also
+specify a name for the combined dataset in the
+Combined dataset text input box.
+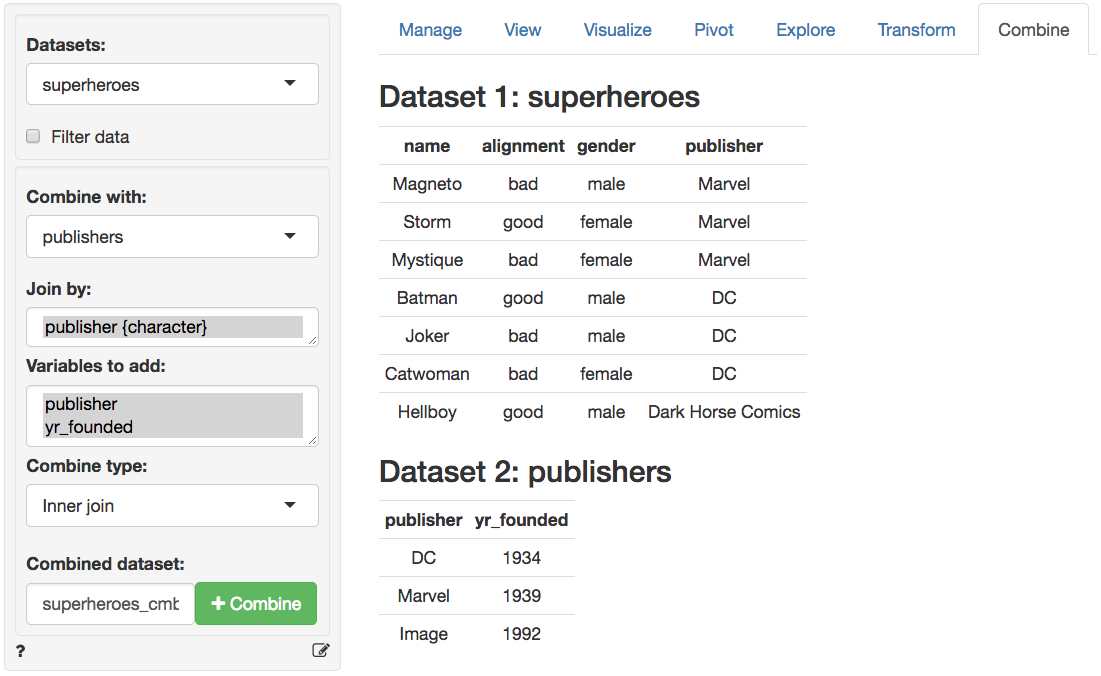
Inner join (superheroes, publishers) +
+If x = superheroes and y = publishers:
+++An inner join returns all rows from x with matching values in y, and +all columns from both x and y. If there are multiple matches between x +and y, all match combinations are returned.
+
| +name + | ++alignment + | ++gender + | ++publisher + | ++yr_founded + | +
|---|---|---|---|---|
| +Magneto + | ++bad + | ++male + | ++Marvel + | ++1939 + | +
| +Storm + | ++good + | ++female + | ++Marvel + | ++1939 + | +
| +Mystique + | ++bad + | ++female + | ++Marvel + | ++1939 + | +
| +Batman + | ++good + | ++male + | ++DC + | ++1934 + | +
| +Joker + | ++bad + | ++male + | ++DC + | ++1934 + | +
| +Catwoman + | ++bad + | ++female + | ++DC + | ++1934 + | +
In the table above we lose Hellboy because, although this
+hero does appear in superheroes, the publisher (Dark
+Horse Comics) does not appear in publishers. The join
+result has all variables from superheroes, plus
+yr_founded, from publishers. We can visualize an
+inner join with the venn-diagram below:
+
The R(adiant) commands are:
+
+# Radiant
+combine_data(superheroes, publishers, by = "publisher", type = "inner_join")
+
+# R
+inner_join(superheroes, publishers, by = "publisher")Left join (superheroes, publishers) +
+++A left join returns all rows from x, and all columns from x and y. If +there are multiple matches between x and y, all match combinations are +returned.
+
| +name + | ++alignment + | ++gender + | ++publisher + | ++yr_founded + | +
|---|---|---|---|---|
| +Magneto + | ++bad + | ++male + | ++Marvel + | ++1939 + | +
| +Storm + | ++good + | ++female + | ++Marvel + | ++1939 + | +
| +Mystique + | ++bad + | ++female + | ++Marvel + | ++1939 + | +
| +Batman + | ++good + | ++male + | ++DC + | ++1934 + | +
| +Joker + | ++bad + | ++male + | ++DC + | ++1934 + | +
| +Catwoman + | ++bad + | ++female + | ++DC + | ++1934 + | +
| +Hellboy + | ++good + | ++male + | ++Dark Horse Comics + | ++NA + | +
The join result contains superheroes with variable
+yr_founded from publishers. Hellboy,
+whose publisher does not appear in publishers, has an
+NA for yr_founded. We can visualize a left join
+with the venn-diagram below:
+
The R(adiant) commands are:
+
+# Radiant
+combine_data(superheroes, publishers, by = "publisher", type = "left_join")
+
+# R
+left_join(superheroes, publishers, by = "publisher")Right join (superheroes, publishers) +
+++A right join returns all rows from y, and all columns from y and x. +If there are multiple matches between y and x, all match combinations +are returned.
+
| +name + | ++alignment + | ++gender + | ++publisher + | ++yr_founded + | +
|---|---|---|---|---|
| +Magneto + | ++bad + | ++male + | ++Marvel + | ++1939 + | +
| +Storm + | ++good + | ++female + | ++Marvel + | ++1939 + | +
| +Mystique + | ++bad + | ++female + | ++Marvel + | ++1939 + | +
| +Batman + | ++good + | ++male + | ++DC + | ++1934 + | +
| +Joker + | ++bad + | ++male + | ++DC + | ++1934 + | +
| +Catwoman + | ++bad + | ++female + | ++DC + | ++1934 + | +
| +NA + | ++NA + | ++NA + | ++Image + | ++1992 + | +
The join result contains all rows and columns from
+publishers and all variables from superheroes.
+We lose Hellboy, whose publisher does not appear in
+publishers. Image is retained in the table but has
+NA values for the variables name,
+alignment, and gender from superheroes.
+Notice that a join can change both the row and variable order so you
+should not rely on these in your analysis. We can visualize a right join
+with the venn-diagram below:
+
The R(adiant) commands are:
+
+# Radiant
+combine_data(superheroes, publishers, by = "publisher", type = "right_join")
+
+# R
+right_join(superheroes, publishers, by = "publisher")Full join (superheroes, publishers) +
+++A full join combines two datasets, keeping rows and columns that +appear in either.
+
| +name + | ++alignment + | ++gender + | ++publisher + | ++yr_founded + | +
|---|---|---|---|---|
| +Magneto + | ++bad + | ++male + | ++Marvel + | ++1939 + | +
| +Storm + | ++good + | ++female + | ++Marvel + | ++1939 + | +
| +Mystique + | ++bad + | ++female + | ++Marvel + | ++1939 + | +
| +Batman + | ++good + | ++male + | ++DC + | ++1934 + | +
| +Joker + | ++bad + | ++male + | ++DC + | ++1934 + | +
| +Catwoman + | ++bad + | ++female + | ++DC + | ++1934 + | +
| +Hellboy + | ++good + | ++male + | ++Dark Horse Comics + | ++NA + | +
| +NA + | ++NA + | ++NA + | ++Image + | ++1992 + | +
In this table we keep Hellboy (even though Dark Horse
+Comics is not in publishers) and Image (even
+though the publisher is not listed in superheroes) and get
+variables from both datasets. Observations without a match are assigned
+the value NA for variables from the other dataset. We can
+visualize a full join with the venn-diagram below:
+
The R(adiant) commands are:
+
+# Radiant
+combine_data(superheroes, publishers, by = "publisher", type = "full_join")
+
+# R
+full_join(superheroes, publishers, by = "publisher")Semi join (superheroes, publishers) +
+++A semi join keeps only columns from x. Whereas an inner join will +return one row of x for each matching row of y, a semi join will never +duplicate rows of x.
+
| +name + | ++alignment + | ++gender + | ++publisher + | +
|---|---|---|---|
| +Magneto + | ++bad + | ++male + | ++Marvel + | +
| +Storm + | ++good + | ++female + | ++Marvel + | +
| +Mystique + | ++bad + | ++female + | ++Marvel + | +
| +Batman + | ++good + | ++male + | ++DC + | +
| +Joker + | ++bad + | ++male + | ++DC + | +
| +Catwoman + | ++bad + | ++female + | ++DC + | +
We get a similar table as with inner_join but it
+contains only the variables in superheroes. The R(adiant)
+commands are:
+# Radiant
+combine_data(superheroes, publishers, by = "publisher", type = "semi_join")
+
+# R
+semi_join(superheroes, publishers, by = "publisher")Anti join (superheroes, publishers) +
+++An anti join returns all rows from x without matching values in y, +keeping only columns from x
+
| +name + | ++alignment + | ++gender + | ++publisher + | +
|---|---|---|---|
| +Hellboy + | ++good + | ++male + | ++Dark Horse Comics + | +
We now get only Hellboy, the only superhero
+not in publishers and we do not get the variable
+yr_founded either. We can visualize an anti join with the
+venn-diagram below:
+
Dataset order +
+Note that the order of the datasets selected may matter for a join. +If we setup the Data > Combine tab as below the results are +as follows:
+
+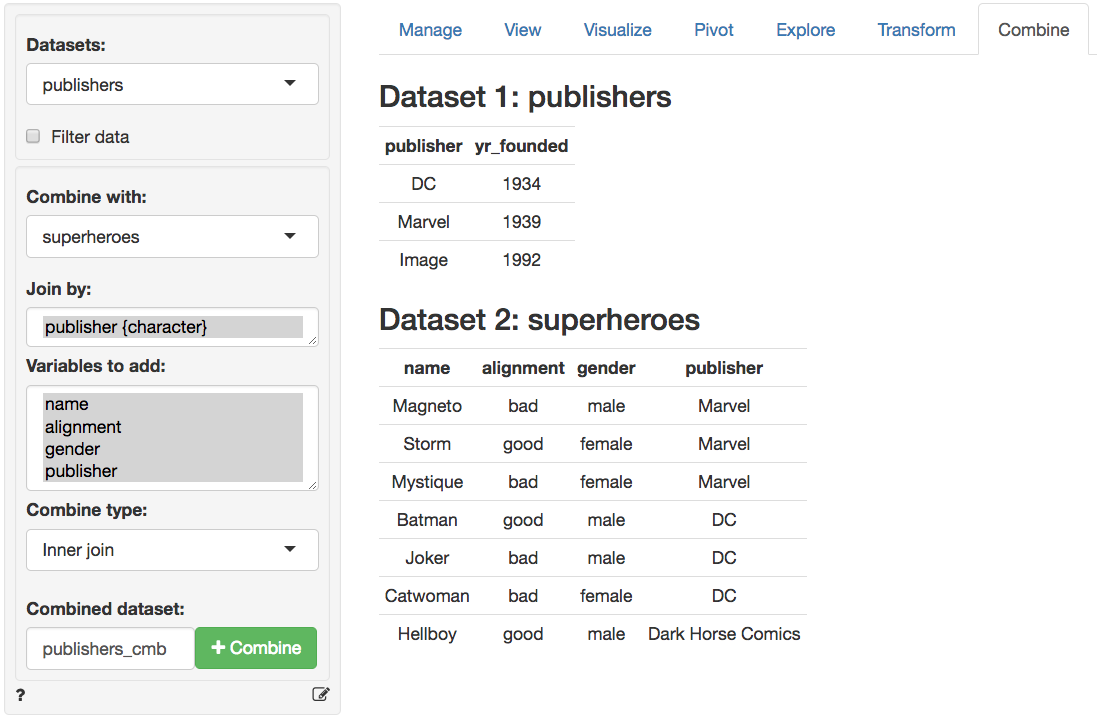
Inner join (publishers, superheroes) +
+| +publisher + | ++yr_founded + | ++name + | ++alignment + | ++gender + | +
|---|---|---|---|---|
| +DC + | ++1934 + | ++Batman + | ++good + | ++male + | +
| +DC + | ++1934 + | ++Joker + | ++bad + | ++male + | +
| +DC + | ++1934 + | ++Catwoman + | ++bad + | ++female + | +
| +Marvel + | ++1939 + | ++Magneto + | ++bad + | ++male + | +
| +Marvel + | ++1939 + | ++Storm + | ++good + | ++female + | +
| +Marvel + | ++1939 + | ++Mystique + | ++bad + | ++female + | +
Every publisher that has a match in superheroes appears
+multiple times, once for each match. Apart from variable and row order,
+this is the same result we had for the inner join shown above.
Left and Right join (publishers, superheroes) +
+Apart from row and variable order, a left join of
+publishers and superheroes is equivalent to a
+right join of superheroes and publishers.
+Similarly, a right join of publishers and
+superheroes is equivalent to a left join of
+superheroes and publishers.
Full join (publishers, superheroes) +
+As you might expect, apart from row and variable order, a full join
+of publishers and superheroes is equivalent to
+a full join of superheroes and publishers.
Semi join (publishers, superheroes) +
+| +publisher + | ++yr_founded + | +
|---|---|
| +DC + | ++1934 + | +
| +Marvel + | ++1939 + | +
With semi join the effect of switching the dataset order is more
+clear. Even though there are multiple matches for each publisher only
+one is shown. Contrast this with an inner join where “If there are
+multiple matches between x and y, all match combinations are returned.”
+We see that publisher Image is lost in the table because it is
+not in superheroes.
Anti join (publishers, superheroes) +
+| +publisher + | ++yr_founded + | +
|---|---|
| +Image + | ++1992 + | +
Only publisher Image is retained because both
+Marvel and DC are in superheroes. We keep
+only variables in publishers.
Additional tools to combine datasets (avengers, superheroes) +
+When two datasets have the same columns (or rows) there are
+additional ways in which we can combine them into a new dataset. We have
+already used the superheroes dataset and will now try to
+combine it with the avengers data. These two datasets have
+the same number of rows and columns and the columns have the same
+names.
In the screen-shot of the Data > Combine tab below we see
+the two datasets. There is no need to select variables to combine the
+datasets here. Any variables in Select variables are
+ignored in the commands below. Again, you can specify a name for the
+combined dataset in the Combined dataset text input
+box.
+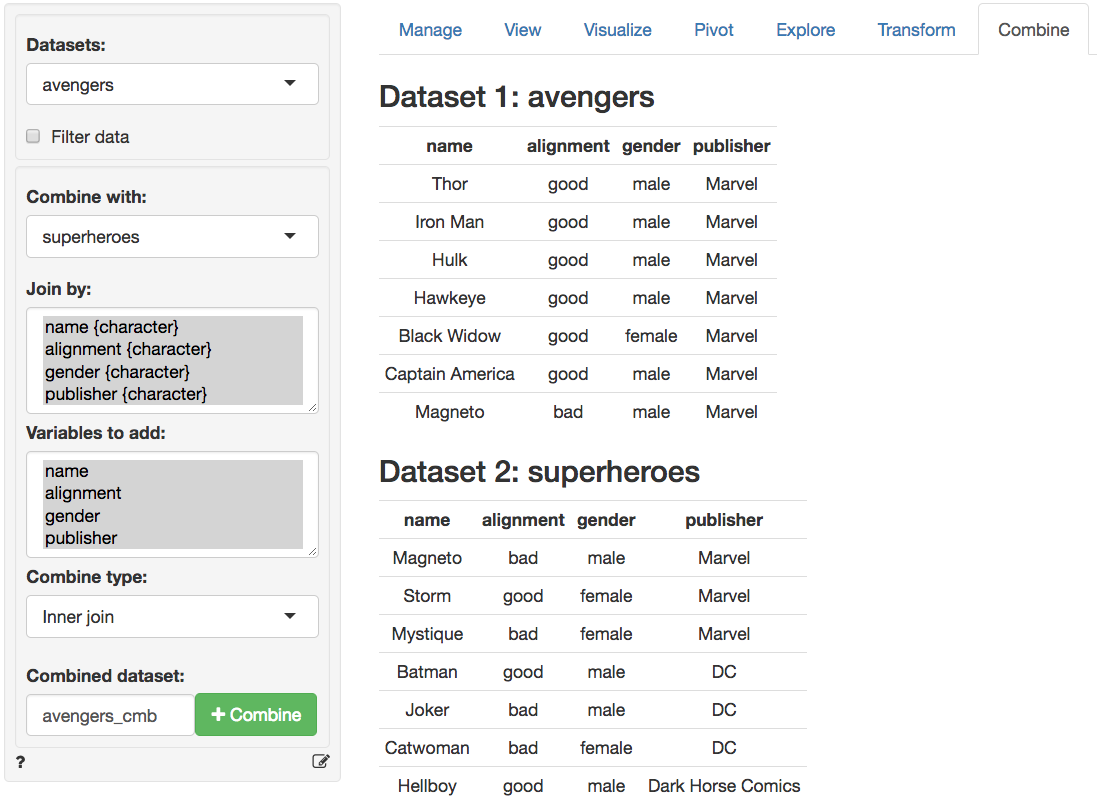
Bind rows +
+| +name + | ++alignment + | ++gender + | ++publisher + | +
|---|---|---|---|
| +Thor + | ++good + | ++male + | ++Marvel + | +
| +Iron Man + | ++good + | ++male + | ++Marvel + | +
| +Hulk + | ++good + | ++male + | ++Marvel + | +
| +Hawkeye + | ++good + | ++male + | ++Marvel + | +
| +Black Widow + | ++good + | ++female + | ++Marvel + | +
| +Captain America + | ++good + | ++male + | ++Marvel + | +
| +Magneto + | ++bad + | ++male + | ++Marvel + | +
| +Magneto + | ++bad + | ++male + | ++Marvel + | +
| +Storm + | ++good + | ++female + | ++Marvel + | +
| +Mystique + | ++bad + | ++female + | ++Marvel + | +
| +Batman + | ++good + | ++male + | ++DC + | +
| +Joker + | ++bad + | ++male + | ++DC + | +
| +Catwoman + | ++bad + | ++female + | ++DC + | +
| +Hellboy + | ++good + | ++male + | ++Dark Horse Comics + | +
If the avengers dataset were meant to extend the list of
+superheroes we could just stack the two datasets, one below the other.
+The new datasets has 14 rows and 4 columns. Due to a coding error in the
+avengers dataset (i.e.., Magneto is not
+an Avenger) there is a duplicate row in the new combined
+dataset. Something we probably don’t want.
The R(adiant) commands are:
+
+# Radiant
+combine_data(avengers, superheroes, type = "bind_rows")
+
+# R
+bind_rows(avengers, superheroes)Bind columns +
+## New names:
+## • `name` -> `name...1`
+## • `alignment` -> `alignment...2`
+## • `gender` -> `gender...3`
+## • `publisher` -> `publisher...4`
+## • `name` -> `name...5`
+## • `alignment` -> `alignment...6`
+## • `gender` -> `gender...7`
+## • `publisher` -> `publisher...8`| +name…1 + | ++alignment…2 + | ++gender…3 + | ++publisher…4 + | ++name…5 + | ++alignment…6 + | ++gender…7 + | ++publisher…8 + | +
|---|---|---|---|---|---|---|---|
| +Thor + | ++good + | ++male + | ++Marvel + | ++Magneto + | ++bad + | ++male + | ++Marvel + | +
| +Iron Man + | ++good + | ++male + | ++Marvel + | ++Storm + | ++good + | ++female + | ++Marvel + | +
| +Hulk + | ++good + | ++male + | ++Marvel + | ++Mystique + | ++bad + | ++female + | ++Marvel + | +
| +Hawkeye + | ++good + | ++male + | ++Marvel + | ++Batman + | ++good + | ++male + | ++DC + | +
| +Black Widow + | ++good + | ++female + | ++Marvel + | ++Joker + | ++bad + | ++male + | ++DC + | +
| +Captain America + | ++good + | ++male + | ++Marvel + | ++Catwoman + | ++bad + | ++female + | ++DC + | +
| +Magneto + | ++bad + | ++male + | ++Marvel + | ++Hellboy + | ++good + | ++male + | ++Dark Horse Comics + | +
If the dataset had different columns for the same superheroes we +could combine the two datasets, side by side. In radiant you will see an +error message if you try to bind these columns because they have the +same name. Something that we should always avoid. The method can be +useful if we know the order of the row ids of two dataset are +the same but the columns are all different.
+Intersect +
+| +name + | ++alignment + | ++gender + | ++publisher + | +
|---|---|---|---|
| +Magneto + | ++bad + | ++male + | ++Marvel + | +
A good way to check if two datasets with the same columns have
+duplicate rows is to choose intersect from the
+Combine type dropdown. There is indeed one row that is
+identical in the avengers and superheroes data
+(i.e., Magneto).
The R(adiant) commands are the same as shown above, except you will
+need to replace bind_rows by intersect.
Union +
+| +name + | ++alignment + | ++gender + | ++publisher + | +
|---|---|---|---|
| +Thor + | ++good + | ++male + | ++Marvel + | +
| +Iron Man + | ++good + | ++male + | ++Marvel + | +
| +Hulk + | ++good + | ++male + | ++Marvel + | +
| +Hawkeye + | ++good + | ++male + | ++Marvel + | +
| +Black Widow + | ++good + | ++female + | ++Marvel + | +
| +Captain America + | ++good + | ++male + | ++Marvel + | +
| +Magneto + | ++bad + | ++male + | ++Marvel + | +
| +Storm + | ++good + | ++female + | ++Marvel + | +
| +Mystique + | ++bad + | ++female + | ++Marvel + | +
| +Batman + | ++good + | ++male + | ++DC + | +
| +Joker + | ++bad + | ++male + | ++DC + | +
| +Catwoman + | ++bad + | ++female + | ++DC + | +
| +Hellboy + | ++good + | ++male + | ++Dark Horse Comics + | +
A union of avengers and
+superheroes will combine the datasets but will omit
+duplicate rows (i.e., it will keep only one copy of the row for
+Magneto). Likely what we want here.
The R(adiant) commands are the same as shown above, except you will
+need to replace bind_rows by union.
Setdiff +
+| +name + | ++alignment + | ++gender + | ++publisher + | +
|---|---|---|---|
| +Thor + | ++good + | ++male + | ++Marvel + | +
| +Iron Man + | ++good + | ++male + | ++Marvel + | +
| +Hulk + | ++good + | ++male + | ++Marvel + | +
| +Hawkeye + | ++good + | ++male + | ++Marvel + | +
| +Black Widow + | ++good + | ++female + | ++Marvel + | +
| +Captain America + | ++good + | ++male + | ++Marvel + | +
Finally, a setdiff will keep rows from
+avengers that are not in superheroes.
+If we reverse the inputs (i.e., choose superheroes from the
+Datasets dropdown and superheroes from the
+Combine with dropdown) we will end up with all rows from
+superheroes that are not in avengers. In both
+cases the entry for Magneto will be omitted.
The R(adiant) commands are the same as shown above, except you will
+need to replace bind_rows by setdiff.
Report > Rmd +
+Add code to
+Report
+> Rmd to (re)create the combined dataset by clicking the
+ icon on the bottom
+left of your screen or by pressing ALT-enter on your
+keyboard.
For additional discussion see the chapter on relational data in +R +for data science and +Tidy +Explain
+R-functions +
+For help with the combine_data function see
+Data
+> Combine
Summarize and explore your data (Data > +Explore)
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/explore.Rmd
+ explore.Rmd++Summarize and explore your data
+
Generate summary statistics for one or more variables in your data. +The most powerful feature in Data > Explore is that you can +easily describe the data by one or more other variables. Where +the +Data +> Pivot tab works best for frequency tables and to summarize +a single numeric variable, the Data > Explore tab allows you +to summarize multiple variables at the same time using various +statistics.
+For example, if we select price from the
+diamonds dataset and click the Create table
+button we can see the number of observations (n), the mean, the
+variance, etc. However, the mean price for each clarity level of the
+diamond can also be easily provided by choosing clarity as
+the Group by variable.
++Note that when a categorical variable (
+factor) is +selected from theNumeric variable(s)dropdown menu it will +be converted to a numeric variable if required for the selected +function. If the factor levels are numeric these will be used in all +calculations. Since the mean, standard deviation, etc. are not relevant +for non-binary categorical variables, these will be converted to 0-1 +(binary) variables where the first level is coded as 1 and all other +levels as 0.
The created summary table can be stored in Radiant by clicking the
+Store button. This can be useful if you want to create
+plots of the summarized data in
+Data
+> Visualize. To download the table to csv format
+click the download icon on the top-right.
You can select options from Column header dropdown to
+switch between different column headers. Select either
+Function (e.g., mean, median, etc), Variable
+(e.g., price, carat, etc), or the levels of the (first)
+Group by variable (e.g., Fair-Ideal).
+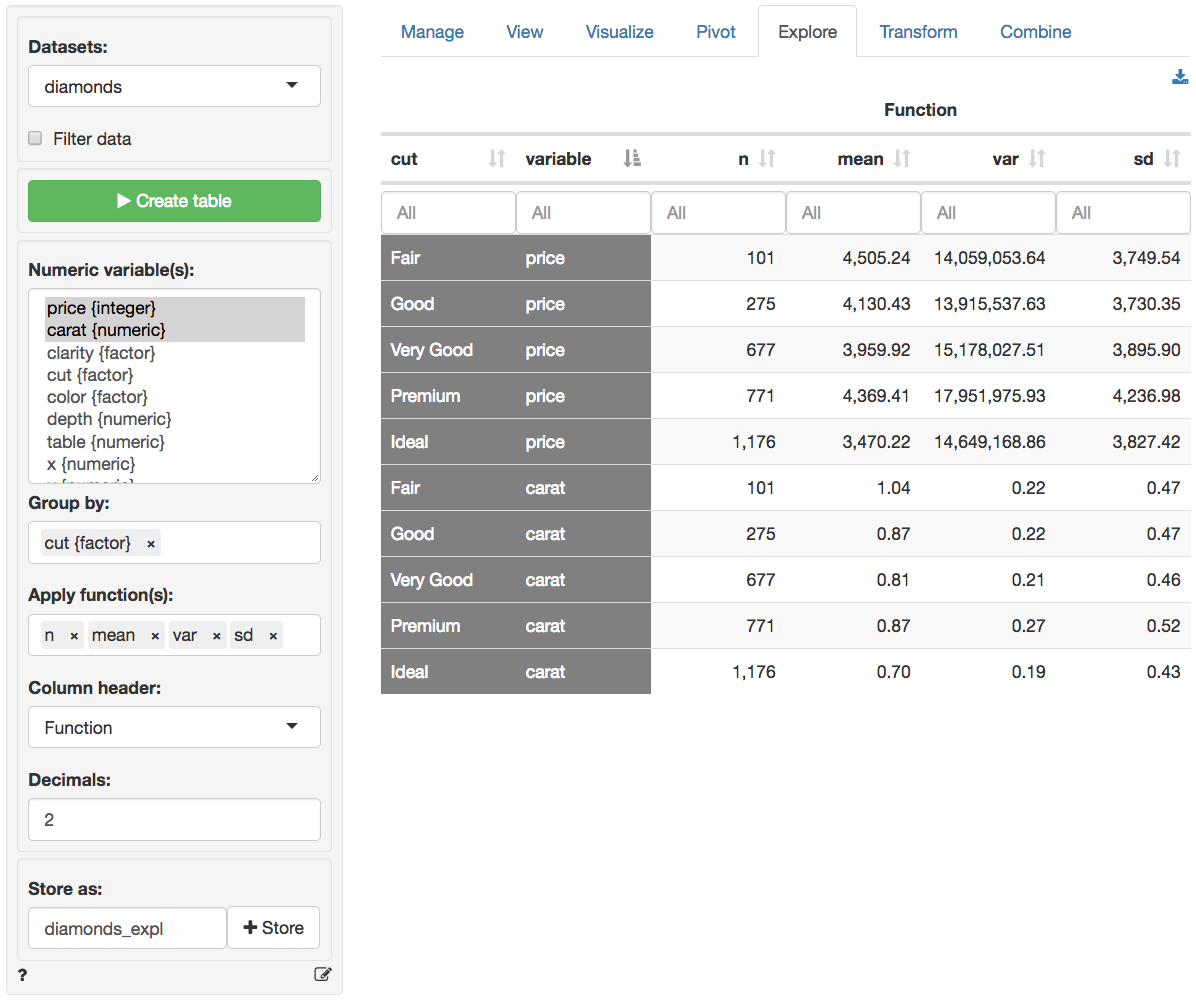
Functions +
+Below you will find a brief description of several functions
+available from the Apply function(s) dropdown menu. Most
+functions, however, will be self-explanatory.
-
+
-
+
ncalculates the number of observations, or rows, in +the data or in a group if aGroup byvariable has been +selected (nuses thelengthfunction in +R)
+ -
+
n_distinctcalculates the number of distinct +values
+ -
+
n_missingcalculates the number of missing values
+ -
+
cvis the coefficient of variation (i.e., mean(x) / +sd(x))
+ -
+
sdandvarcalculate the sample standard +deviation and variance for numeric data
+ -
+
mecalculates the margin of error for a numeric +variable using a 95% confidence level
+ -
+
propcalculates a proportion. For a variable with only +values 0 or 1 this is equivalent tomean. For other numeric +variables it captures the occurrence of the maximum value. For a +factorit captures the occurrence of the first level.
+ -
+
sdpropandvarpropcalculate the sample +standard deviation and variance for a proportion
+ -
+
mepropcalculates the margin of error for a proportion +using a 95% confidence level
+ -
+
sdpopandvarpopcalculate the population +standard deviation and variance
+
Filter data +
+Use the Filter data box to select (or omit) specific
+sets of rows from the data. See the helpfile for
+Data
+> View for details.
Report > Rmd +
+Add code to
+Report
+> Rmd to (re)create the summary table by clicking the
+ icon on the bottom
+left of your screen or by pressing ALT-enter on your
+keyboard.
R-functions +
+For an overview of related R-functions used by Radiant to summarize +and explore data see +Data +> Explore
+ +Loading and Saving data (Data > Manage)
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/manage.Rmd
+ manage.Rmd++Manage data and state: Load data into Radiant, Save data to disk, +Remove a dataset from memory, or Save/Load the state of the app
+
Datasets +
+When you first start Radiant a dataset (diamonds) with
+information on diamond prices is shown.
It is good practice to add a description of the data and variables to
+each file you use. For the files that are bundled with Radiant you will
+see a brief overview of the variables etc. below a table of the first 10
+rows of the data. To add a description for your own data click the
+Add/edit data description check-box. A text-input box will
+open below the table where you can add text in
+markdown
+format. The description provided for the diamonds data
+included with Radiant should serve as a good example. After adding or
+editing a description click the Update description
+button.
To rename a dataset loaded in Radiant click the
+Rename data check box, enter a new name, and click the
+Rename button
Load data +
+The best way to load and save data for use in Radiant (and R) is to
+use the R-data format (rds or rda). These are binary files that can be
+stored compactly and read into R quickly. Select rds (or
+rda) from the Load data of type dropdown and
+click Browse to locate the file(s) you want to load on your
+computer.
You can get data from a spreadsheet (e.g., Excel or Google sheets)
+into Radiant in two ways. First, you can save data from the spreadsheet
+in csv format and then, in Radiant, choose csv from the
+Load data of type dropdown. Most likely you will have a
+header row in the csv file with variable names. If the data are not
+comma separated you can choose semicolon or tab separated. To load a csv
+file click ‘Browse’ and locate the file on your computer.
Alternatively, you can select and copy the data in the spreadsheet
+using CTRL-C (or CMD-C on mac), go to Radiant, choose
+clipboard from the Load data of type dropdown,
+and click the Paste button. This is a short-cut that can be
+convenient for smaller datasets that are cleanly formatted.
If the data is available in R’s global workspace (e.g., you opened a
+data set in Rstudio and then started Radiant from the
+addins menu) you can move (or copy) it to Radiant by
+selecting from global workspace. Select the data.frame(s)
+you want to use and click the Load button.
To access all data files bundled with Radiant choose
+examples from the Load data of type dropdown
+and then click the Load button. These files are used to
+illustrate the various data and analysis tools accessible in Radiant.
+For example, the avengers and publishers data
+are used to illustrate how to combine data in R(adiant) (i.e., Data
+> Combine).
If csv data is available online choose
+csv (url) from the dropdown, paste the url into the text
+input shown, and press Load. If an rda file is
+available online choose rda (url) from the dropdown, paste
+the url into the text input, and press Load.
Save data +
+As mentioned above, the most convenient way to get data in and out of
+Radiant is to use the R-data format (rds or rda). Choose
+rds (or rda) from the
+Save data to type dropdown and click the Save
+button to save the selected dataset to file.
Again, it is good practice to add a description of the data and
+variables to each file you use. To add a description for your own data
+click the ‘Add/edit data description’ check-box, add text to the
+text-input window shown in
+markdown
+format, and then click the Update description button. When
+you save the data as an rds (or rda) file the description you created
+(or edited) will automatically be added to the file as an
+attribute.
Getting data from Radiant into a spreadsheet can be achieved in two
+ways. First, you can save data in csv format and load the file into the
+spreadsheet (i.e., choose csv from the
+Save data to type dropdown and click the Save
+button). Alternatively, you can copy the data from Radiant into the
+clipboard by choosing clipboard from the dropdown and
+clicking the Copy button, open the spreadsheet, and paste
+the data from Radiant using CTRL-V (or CMD-V on mac).
To move or copy data from Radiant into R(studio)’s global workspace
+select to global workspace from the
+Save data to type dropdown and click the Save
+button.
Save and load state +
+It is convenient to work with state files if you want complete your
+work at another time, perhaps on another computer, or to review previous
+work you completed using Radiant. You can save and load the state of the
+Radiant app just as you would a data file. The state file (extension
+.state.rda) will contain (1) the data loaded in Radiant,
+(2) settings for the analyses you were working on, (3) and any reports
+or code from the Report menu. To save the current state of the
+app to your hard-disk click the
+icon in the navbar and then click Save radiant state file.
+To load load a previous state click the
+ icon in the navbar and the click
+Load radiant state file.
You can also share a state file with others that would like to
+replicate your analyses. As an example, download and then load the state
+file
+radiant-example.state.rda
+as described above. You will navigate automatically to the Data >
+Visualize tab and will see a plot. See also the Data >
+View tab for some additional settings loaded from the state file.
+There is also a report in Report > Rmd created using the
+Radiant interface. The html file
+radiant-example.nb.html
+contains the output created by clicking the Knit report
+button.
Loading and saving state also works with Rstudio. If you start
+Radiant from Rstudio and use
+ and then click
+Stop, the r_data environment and the
+r_info and r_state lists will be put into
+Rstudio’s global workspace. If you start radiant again from the
+Addins menu it will use r_data,
+r_info, and r_state to restore state. Also, if
+you load a state file directly into Rstudio it will be used when you
+start Radiant.
Use Refresh in the
+ menu in the navbar to
+return to a clean/new state.
Remove data from memory +
+If data are loaded in memory that you no longer need in the current
+session check the Remove data from memory box. Then select
+the data to remove and click the Remove data button. One
+datafile will always remain open.
Using commands to load and save data +
+R-code can be used in Report > Rmd or Report >
+R to load data from a file directly into the active Radiant
+session. Use register("insert-dataset-name") to add a
+dataset to the Datasets dropdown. R-code can also be used
+to extract data from Radiant and save it to disk.
R-functions +
+For an overview of related R-functions used by Radiant to load and +save data see +Data +> Manage
+ +Create pivot tables (Data > Pivot)
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/pivotr.Rmd
+ pivotr.Rmd++Create pivot tables to explore your data
+
If you have used pivot-tables in Excel the functionality provided in +the Data > Pivot tab should be familiar to you. Similar to +the +Data +> Explore tab, you can generate summary statistics for +variables in your data. You can also generate frequency tables. Perhaps +the most powerful feature in Data > Pivot is that you can +easily describe the data by one or more other variables.
+For example, with the diamonds data loaded, select
+clarity and cut from the
+Categorical variables drop-down. The categories for the
+first variable will be the column headers but you can drag-and-drop the
+selected variables to change their ordering. After selecting these two
+variables, and clicking on the Create pivot table button, a
+frequency table of diamonds with different levels of clarity and quality
+of cut is shown. Choose Row, Column, or
+Total from the Normalize by drop-down to
+normalize cell frequencies or create an index from a summary statistic
+by the row, column, or overall total. If a normalize option is selected
+it can be convenient to check the Percentage box to express
+the numbers as percentages. Choose Color bar or
+Heat map from the Conditional formatting
+drop-down to emphasize the highest frequency counts.
It is also possible to summarize numerical variables. Select
+price from the Numeric variables drop-down.
+This will create the table shown below. Just as in the
+Data
+> View tab you can sort the table by clicking on the column
+headers. You can also use sliders (e.g., click in the input box below
+I1) to limit the view to values in a specified range. To
+view only information for diamonds with a Very good,
+Premium or Ideal cut click in the input box
+below the cut header.
+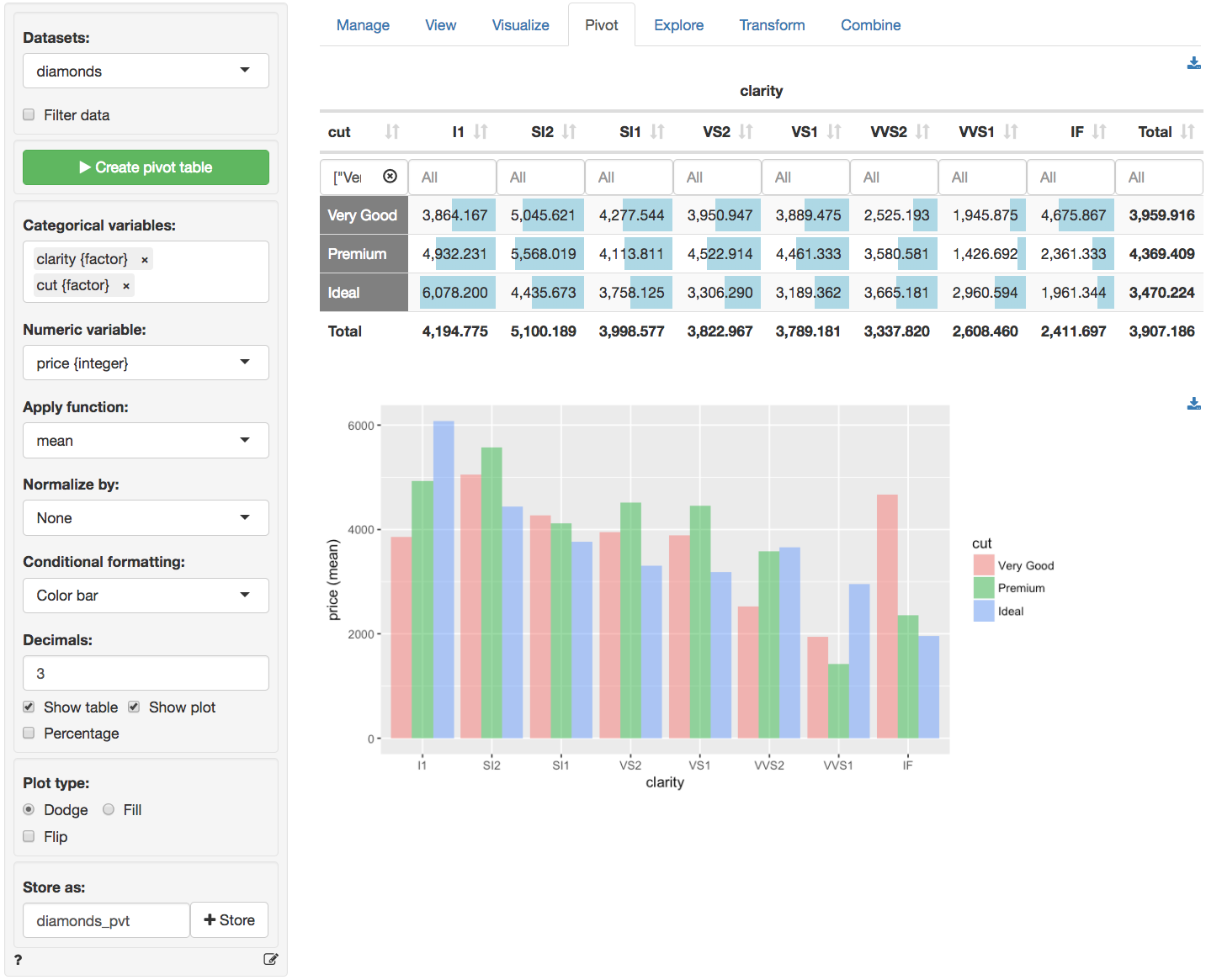
Below you will find a brief description of several functions
+available from the Apply function dropdown menu. Most
+functions, however, will be self-explanatory.
-
+
-
+
ncalculates the number of observations, or rows, in +the data or in a group if aGroup byvariable has been +selected (nuses thelengthfunction in +R)
+ -
+
n_distinctcalculates the number of distinct +values
+ -
+
n_missingcalculates the number of missing values
+ -
+
cvis the coefficient of variation (i.e., mean(x) / +sd(x))
+ -
+
sdandvarcalculate the sample standard +deviation and variance for numeric data
+ -
+
mecalculates the margin of error for a numeric +variable using a 95% confidence level
+ -
+
propcalculates a proportion. For a variable with only +values 0 or 1 this is equivalent tomean. For other numeric +variables it captures the occurrence of the maximum value. For a +factorit captures the occurrence of the first level.
+ -
+
sdpropandvarpropcalculate the sample +standard deviation and variance for a proportion
+ -
+
mepropcalculates the margin of error for a proportion +using a 95% confidence level
+ -
+
sdpopandvarpopcalculate the population +standard deviation and variance
+
You can also create a bar chart based on the generated table (see +image above). To download the table in csv format or the plot +in png format click the appropriate download icon on the +right.
+++Note that when a categorical variable (
+factor) is +selected from theNumeric variable(s)dropdown menu it will +be converted to a numeric variable if required for the selected +function(s). If the factor levels are numeric these will be used in all +calculations. Since the mean, standard deviation, etc. are not relevant +for non-binary categorical variables, these will be converted to 0-1 +(binary) variables where the first level is coded as 1 and all other +levels as 0.
Filter data +
+Use the Filter data box to select (or omit) specific
+sets of rows from the data to tabulate. See the help file for
+Data
+> View for details.
Store +
+The created pivot table can be stored in Radiant by clicking the
+Store button. This can be useful if you want do additional
+analysis on the table or to create plots of the summarized data in
+Data
+> Visualize. To download the table to csv format
+click the download icon on the top-right.
Report > Rmd +
+Add code to
+Report
+> Rmd to (re)create the pivot table by clicking the
+ icon on the bottom
+left of your screen or by pressing ALT-enter on your
+keyboard.
If a plot was created it can be customized using ggplot2
+commands (e.g.,
+plot(result) + labs(title = "Pivot graph")). See
+Data
+> Visualize for details.
R-functions +
+For an overview of related R-functions used by Radiant to create +pivot tables see +Data +> Pivot
+ +Create a reproducible report using R (Report +> R)
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/report_r.Rmd
+ report_r.Rmd++Create a (reproducible) report using R
+
The Report > R tab allows you to run R-code with access
+to all functions and data in Radiant. By clicking the
+Knit report (R) button, the code will be evaluated and the
+output will be shown on the right of the Report > R page. To
+evaluate only a part of the code use the cursor to select a section and
+press CTRL-enter (CMD-enter on mac).
You can load an R-code file into Radiant by clicking the
+Load report button and selecting an .r or .R file. If you
+started Radiant from Rstudio you can save a report in HTML, Word, or PDF
+format by selecting the desired format from the drop-down menu and
+clicking Save report. To save just the code choose
+R from the dropdown and press the Save report
+button.
If you started Radiant from Rstudio, you can also click the
+Read files button to browse for files and generate code to
+read it into Radiant. For example, read rda, rds, xls, yaml, and feather
+and add them to the Datasets dropdown. If the file type you
+want to load is not currently supported, the path to the file will be
+returned. The file path used will be relative to the Rstudio-project
+root. Paths to files synced to a local Dropbox or Google Drive folder
+will use the find_dropbox and find_gdrive
+functions to enhances reproducibility.
As an example you can copy-and-paste the code below into the editor
+and press Knit report (R) to generate results.
+## get the active dataset and show the first few observations
+.get_data() %>%
+ head()
+
+## access a dataset
+diamonds %>%
+ select(price, clarity) %>%
+ head()
+
+## add a variable to the diamonds data
+diamonds <- mutate(diamonds, log_price = log(price))
+
+## show the first observations in the price and log_price columns
+diamonds %>%
+ select(price, log_price) %>%
+ head()
+
+## create a histogram of prices
+diamonds %>%
+ ggplot(aes(x = price)) +
+ geom_histogram()
+
+## and a histogram of log-prices using radiant.data::visualize
+visualize(diamonds, xvar = "log_price", custom = TRUE)
+
+## open help in the R-studio viewer from Radiant
+help(package = "radiant.data")
+
+## If you are familiar with Shiny you can call reactives when the code
+## is evaluated inside a Shiny app. For example, if you transformed
+## some variables in Data > Transform you can call the transform_main
+## reacive to see the latest result. Very useful for debugging
+# transform_main() %>% head()
+head()Options +
+The editor used in Report > Rmd and Report >
+R has several options that can be set in
+.Rprofile.
+options(radiant.ace_vim.keys = FALSE)
+options(radiant.ace_theme = "cobalt")
+options(radiant.ace_tabSize = 2)
+options(radiant.ace_useSoftTabs = TRUE)
+options(radiant.ace_showInvisibles = TRUE)
+options(radiant.ace_autoComplete = "live")Notes:
+-
+
-
+
vim.keysenables a set of special keyboard short-cuts. +If you have never used VIM you probably don’t want this
+ - For an overview of available editor themes see:
+
shinyAce::getAceThemes()+
+ - Tabs are converted to 2 spaces by default (i.e., ‘soft’ tabs). You +can change the number of spaces used from 2 to, for example, 4 +
-
+
showInvisiblesshows tabs and spaces in the editor
+ - Autocomplete has options “live”, “enabled”, and “disabled” +
R-functions +
+For an overview of related R-functions used by Radiant to generate +reproducible reports see +Report
+ +Create a reproducible report using Rmarkdown +(Report > Rmd)
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/report_rmd.Rmd
+ report_rmd.Rmd++Create a (reproducible) report using Rmarkdown
+
The best way to store your work in Radiant is to use the Report +> Rmd feature and save a state file with all your results and +settings. The report feature in Radiant should be used in conjunction +with the icons shown +on the bottom left of your screen on most pages.
+The editor shown on the left in Report > Rmd shows past
+commands in R-code chunks. These chunks can
+include R-code you typed or R-code generated by Radiant and added to the
+report after clicking an
+ icon. All code chunks
+start with ```{r} and are closed by ```
By default Radiant will add the R-code generated for the analysis you +just completed to the bottom of the report. After clicking a + icon Radiant will, by +default, switch to the Report > Rmd tab. Click inside the +editor window on the left and scroll down to see the generated +commands.
+If you want more control over where the R-code generated by Radiant
+is put into your report, choose Manual paste instead or
+Auto paste from the appropriate drop-down in the Report
+> Rmd tab. When Manual paste is selected, the code
+is put into the clipboard when you click
+ and you can paste it
+where you want in the editor window.
If you started Radiant from Rstudio, you can also choose to have
+commands sent to an Rmarkdown (R-code) document open in Rstudio by
+selecting To Rmd (To R) instead of
+Auto paste or Manual paste. If you choose
+To Rmd the editor in Report > Rmd will be
+hidden (i.e., “Preview only”) and clicking on
+Knit report (Rmd) will source the text and code directly
+from Rstudio.
By default, the app will switch to the Report > Rmd tab
+after you click the
+icon. However, if you don’t want to switch tabs after clicking a
+ icon, choose
+Don't switch tab from the appropriate drop-down in the
+Report > Rmd tab. Don't switch tab is the
+default option when you choose To Rmd.
You can add text or additional commands to create an Rmarkdown +document. An Rmarkdown file (extension .Rmd) is a plain text file that +can be opened in Notepad (Windows), TextEdit (Mac), Rstudio, Sublime +Text, or any other text editor. Please do not use Word +to edit Rmarkdown files.
+Using Rmarkdown is extremely powerful because you can replicate your
+entire analysis quickly without having to generate all the required
+R-code again. By clicking the Knit report (Rmd) button on
+the top-left of your screen, the output from the analysis will be
+(re)created and shown on the right of the Report > Rmd page.
+To evaluate only a part of the report use the cursor to select a section
+and press CTRL-enter (CMD-enter on mac) to
+create the (partial) output.
You can add text, bullets, headers, etc. around the code chunks to +describe and explain the results using +markdown. +For an interactive markdown tutorial visit +commonmark.org.
+If you started Radiant from Rstudio you can save the report in
+various formats (i.e., Notebook, HTML, Word, Powerpoint, or PDF). For
+more on generating powerpoint presentation see
+https://bookdown.org/yihui/rmarkdown/powerpoint-presentation.html.
+To save the Rmarkdown file open in the editor select Rmd
+(or Rmd + Data (zip)) and press Save report.
+Previously saved Rmarkdown files can be loaded into Radiant by using the
+Load report button. For more
You can also click the Read files button to browse for
+files and generate code to read it into Radiant. For example, read rda,
+rds, xls, yaml, and feather and add them to the Datasets
+dropdown. You can also read images, R-code, and text (e.g., Rmd or md)
+to include in your report. If the file type you want to load is not
+currently supported, the path to the file will be returned. If Radiant
+was started from an Rstudio project, the file paths used will be
+relative to the project root. Paths to files synced to local Dropbox or
+Google Drive folder will use the find_dropbox and
+find_gdrive functions to enhances reproducibility.
State +
+The best way to save your analyses and settings is to save the
+state of the application to a file by clicking on the
+ icon in the navbar and then
+clicking on Save radiant state file. The state file
+(extension rda) will contain (1) the data loaded in Radiant, (2)
+settings for the analyses you were working on, (3) and any reports or
+code from the Report > Rmd and Report > R. Save
+the state file to your hard-disk and, when you are ready to continue,
+simply load it by icon in the
+navbar and then clicking on Load radiant state file
If you are using Radiant for a class I suggest you use the Report
+> Rmd feature to complete assignments and cases. When you are
+done, generate an (HTML) Notebook (or Word or PDF) report by clicking
+the Save report button. Submit both the report and your
+state file.
Options +
+The editor used in Report > Rmd and Report >
+R has several options that can be set in .Rprofile.
+You can use usethis::edit_r_profile() to alter the settings
+in .Rprofile
+options(radiant.ace_vim.keys = FALSE)
+options(radiant.ace_theme = "cobalt")
+options(radiant.ace_tabSize = 2)
+options(radiant.ace_useSoftTabs = TRUE)
+options(radiant.ace_showInvisibles = TRUE)
+options(radiant.ace_autoComplete = "live")
+options(radiant.powerpoint_style = "~/Dropbox/rmd-styles/style.potx")
+options(radiant.word_style = "~/Dropbox/rmd-styles/style.docx")Notes:
+-
+
-
+
vim.keysenables a set of special keyboard short-cuts. +If you have never used VIM you probably don’t want this
+ - For an overview of available editor themes see:
+
shinyAce::getAceThemes()+
+ - Tabs are converted to 2 spaces by default (i.e., ‘soft’ tabs). You +can change the number of spaces used from 2 to, for example, 4 +
-
+
showInvisiblesshows tabs and spaces in the editor
+ - Autocomplete has options “live”, “enabled”, and “disabled” +
- Radiant has default styles for Word and Powerpoint files. These can
+be replaced with styles files you created however. Click the links below
+to download the style files used in Radiant to your computer. Edit the
+files and use
optionsas shown above to tell Radiant where +to find the style files you want to use. + +
+
R-functions +
+For an overview of related R-functions used by Radiant to generate +reproducible reports see +Report
+ +Loading and Saving the State of the +application
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/state.Rmd
+ state.Rmd++Save, load, share, or view state
+
It is convenient to work with state files if you want complete your
+work at another time, perhaps on another computer, or to review previous
+work you completed using Radiant. You can save and load the state of the
+Radiant app just as you would a data file. The state file (extension
+.rda) will contain (1) the data loaded in Radiant, (2)
+settings for the analyses you were working on, (3) and any reports or
+code from the Report menu. To save the current state of the app
+to your hard-disk click the icon
+in the navbar and then click Save radiant state file. To
+load load a previous state click the
+ icon in the navbar and the click
+Load radiant state file.
You can also share a state file with others that would like to
+replicate your analyses. As an example, download and then load the state
+file
+radiant-example.state.rda
+as described above. You will navigate automatically to the Data >
+Visualize tab and will see a plot. See also the Data >
+View tab for some additional settings loaded from the state file.
+There is also a report in Report > Rmd created using the
+Radiant interface. The html file
+radiant-example.nb.html
+contains the output created by clicking the Knit report
+button.
Loading and saving state also works with Rstudio. If you start
+Radiant from Rstudio and use
+ and then click
+Stop, the r_data environment and the
+r_info and r_state lists will be put into
+Rstudio’s global workspace. If you start radiant again from the
+Addins menu it will use r_data,
+r_info, and r_state to restore state. Also, if
+you load a state file directly into Rstudio it will be used when you
+start Radiant.
Use Refresh in the
+ menu in the navbar to
+return to a clean/new state.
Transform variables (Data > Transform)
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/transform.Rmd
+ transform.Rmd++Transform variables
+
Transform command log +
+All transformations applied in the Data > Transform tab
+can be logged. If, for example, you apply a
+Ln (natural log) transformation to numeric variables the
+following code is generated and put in the
+Transform command log window at the bottom of your screen
+when you click the Store button.
+## transform variable
+diamonds <- mutate_ext(
+ diamonds,
+ .vars = vars(price, carat),
+ .funs = log,
+ .ext = "_ln"
+)This is an important feature if you want to re-run a report with new, +but similar, data. Even more important is that there is a record of the +steps taken to transform the data and to generate results, i.e., your +work is now reproducible.
+To add commands contained in the command log window to a report in +Report +> Rmd click the + icon.
+Filter data +
+Even if a filter has been specified it will be ignored for (most)
+functions available in Data > Transform. To create a new
+dataset based on a filter navigate to the
+Data
+> View tab and click the Store button.
+Alternatively, to create a new dataset based on a filter, select
+Split data > Holdout sample from the
+Transformation type dropdown.
Hide summaries +
+For larger datasets, or when summaries are not needed, it can useful
+to click Hide summariesbefore selecting the transformation
+type and specifying how you want to alter the data. If you do want to
+see summaries make sure that Hide summaries is not
+checked.
Change variables +
+Bin +
+The Bin command is a convenience function for the
+xtile command discussed below when you want to create
+multiple quintile/decile/… variables. To calculate quintiles enter
+5 as the Nr bins. The reverse
+option replaces 1 by 5, 2 by 4, …, 5 by 1. Choose an appropriate
+extension for the new variable(s).
Change type +
+When you select Type from the
+Transformation type drop-down another drop-down menu is
+shown that will allow you to change the type (or class) of one or more
+variables. For example, you can change a variable of type integer to a
+variable of type factor. Click the Store button to commit
+the changes to the data set. A description of the transformation options
+is provided below.
-
+
- As factor: convert a variable to type factor (i.e., a categorical +variable) +
- As number: convert a variable to type numeric +
- As integer: convert a variable to type integer +
- As character: convert a variable to type character (i.e., +strings) +
- As times series: convert a variable to type ts +
- As date (mdy): convert a variable to a date if the dates are +structured as month-day-year +
- As date (dmy): convert a variable to a date if the dates are +structured as day-month-year +
- As date (ymd): convert a variable to a date if the dates are +structured as year-month-day +
- As date/time (mdy_hms): convert a variable to a date if the dates +are structured as month-day-year-hour-minute-second +
- As date/time (mdy_hm): convert a variable to a date if the dates are +structured as month-day-year-hour-minute +
- As date/time (dmy_hms): convert a variable to a date if the dates +are structured as day-month-year-hour-minute-second +
- As date/time (dmy_hm): convert a variable to a date if the dates are +structured as day-month-year-hour-minute +
- As date/time (ymd_hms): convert a variable to a date if the dates +are structured as year-month-day-hour-minute-second +
- As date/time (ymd_hm): convert a variable to a date if the dates are +structured as year-month-day-hour-minute +
Note: When converting a variable to type
+ts (i.e., time series) you should, at least, specify a
+starting period and the frequency data. For example, for weekly data
+that starts in the 4th week of the year, enter 4 as the
+Start period and set Frequency to
+52.
Normalize +
+Choose Normalize from the
+Transformation type drop-down to standardize one or more
+variables. For example, in the diamonds data we may want to express
+price of a diamond per-carat. Select carat as the
+Normalizing variable and price in the
+Select variable(s) box. You will see summary statistics for
+the new variable (e.g., price_carat) in the main panel.
+Commit changes to the data by clicking the Store
+button.
Recode +
+To use the recode feature select the variable you want to change and
+choose Recode from the Transformation type
+drop-down. Provide one or more recode commands, separated by a
+;, and press return to see information about the changed
+variable. Note that you can specify a name for the recoded variable in
+the Recoded variable name input box (press return to submit
+changes). Finally, click Store to add the recoded variable
+to the data. Some examples are given below.
-
+
- Set values below 20 to
Lowand all others to +High+
+
-
+
- Set above 20 to
Highand all others to +Low+
+
-
+
- Set values 1 through 12 to
A, 13:24 toB, +and the remainder toC+
+
-
+
- Collapse age categories for a
+Basics
+> Tables > Cross-tabs cross-tab analysis. In the example
+below
<25and25-34are recoded to +<35,35-44and35-44are +recoded to35-54, and55-64and +>64are recoded to>54+
+
+'<25' = '<35'; '25-34' = '<35'; '35-44' = '35-54'; '45-54' = '35-54'; '55-64' = '>54'; '>64' = '>54'-
+
- To exclude a particular value (e.g., an outlier in the data) for
+subsequent analyses we can recode it to a missing value. For example, if
+we want to remove the maximum value from a variable called
+
salesthat is equal to 400 we would (1) select the variable +salesin theSelect variable(s)box and enter +the command below in theRecodebox. Press +returnandStoreto add the recoded variable +to the data
+
+400 = NA-
+
- To recode specific numeric values (e.g., carat) to a new value (1)
+select the variable
caratin the +Select variable(s)box and enter the command below in the +Recodebox to set the value for carat to 2 in all rows +where carat is currently larger than or equal to 2. Press +returnandStoreto add the recoded variable +to the data
+
+2:hi = 2Note: Do not use = in a variable label
+when using the recode function (e.g., 50:hi = '>= 50')
+as this will cause an error.
Reorder or remove levels +
+If a (single) variable of type factor is selected in
+Select variable(s), choose
+Reorder/Remove levels from the
+Transformation type drop-down to reorder and/or remove
+levels. Drag-and-drop levels to reorder them or click the \(\times\) to remove them. Note that, by
+default, removing one or more levels will introduce missing values in
+the data. If you prefer to recode the removed levels into a new level,
+for example “other”, simply type “other” in the
+Replacement level name input box and press
+return. If the resulting factor levels appear as intended,
+press Store to commit the changes. To temporarily exclude
+levels from the data use the Filter data box (see the help
+file linked in the
+Data
+> View tab).
Rename +
+Choose Rename from the Transformation type
+drop-down, select one or more variables, and enter new names for them in
+the Rename box. Separate names by a ,. Press
+return to see summaries for the renamed variables on screen and press
+Store to alter the variable names in the data.
Replace +
+Choose Replace from the Transformation type
+drop-down if you want to replace existing variables in the data with new
+ones created using, for example, Create,
+Transform, Clipboard, etc.. Select one or more
+variables to overwrite and the same number of replacement variables.
+Press Store to alter the data.
Transform +
+When you select Transform from the
+Transformation type drop-down another drop-down menu is
+shown you can use to apply common transformations to one or more
+variables in the data. For example, to take the (natural) log of a
+variable select the variable(s) you want to transform and choose
+Ln (natural log) from the Apply function
+drop-down. The transformed variable will have the extension specified in
+the Variable name extension input (e.g,. _ln).
+Make sure to press return after changing the extension.
+Click the Store button to add the (changed) variable(s) to
+the data set. A description of the transformation functions included in
+Radiant is provided below.
-
+
- Ln: create a natural log-transformed version of the selected +variable (i.e., log(x) or ln(x)) +
- Square: multiply a variable by itself (i.e., x^2 or square(x)) +
- Square-root: take the square-root of a variable (i.e., x^.5) +
- Absolute: Absolute value of a variable (i.e., abs(x)) +
- Center: create a new variable with a mean of zero (i.e., x - +mean(x)) +
- Standardize: create a new variable with a mean of zero and standard +deviation of one (i.e., (x - mean(x))/sd(x)) +
- Inverse: 1/x +
Create new variable(s) +
+Clipboard +
+Although not recommended, you can manipulate your data in a
+spreadsheet (e.g., Excel or Google sheets) and copy-and-paste the data
+back into Radiant. If you don’t have the original data in a spreadsheet
+already use the clipboard feature in
+Data
+> Manage so you can paste it into the spreadsheet or click
+the download icon on the top right of your screen in the
+Data
+> View tab. Apply your transformations in the spreadsheet
+program and then copy the new variable(s), with a header label, to the
+clipboard (i.e., CTRL-C on windows and CMD-C on mac). Select
+Clipboard from the Transformation type
+drop-down and paste the new data into the
+Paste from spreadsheet box. It is key that new variable(s)
+have the same number of observations as the data in Radiant. To add the
+new variables to the data click Store.
++Note: Using the clipboard feature for data +transformation is discouraged because it is not reproducible.
+
Create +
+Choose Create from the Transformation type
+drop-down. This is the most flexible command to create new or transform
+existing variables. However, it also requires some basic knowledge of
+R-syntax. A new variable can be any function of other variables in the
+(active) dataset. Some examples are given below. In each example the
+name to the left of the = sign is the name of the new
+variable. To the right of the = sign you can include other
+variable names and basic R-functions. After you type the command press
+return to see summary statistics for the new variable. If
+the result is as expected press Store to add it to the
+dataset.
++Note: If one or more variables is selected from the +
+Select variableslist they will be used to group +the data before creating the new variable (see example 1. below). If +this is not the intended result make sure that no variables are selected +when creating new variables
-
+
- Create a new variable
zthat is equal to the mean of +price. To calculate the mean of price per group (e.g., per level of +clarity) selectclarityfrom the +Select variableslist before creatingz+
+
+z = mean(price)-
+
- Create a new variable
zthat is the difference between +variables x and y
+
+z = x - y-
+
- Create a new variable
zthat is a transformation of +variablexwith mean equal to zero (see also +Transform > Center):
+
+z = x - mean(x)-
+
- Create a new _logical) variable
zthat takes on the +value TRUE whenx > yand FALSE otherwise
+
+z = x > y-
+
- Create a new logical
zthat takes on the value +TRUE whenxis equal toyand FALSE +otherwise
+
+z = x == y-
+
- Create a variable
zthat is equal tox+lagged by 3 periods
+
+z = lag(x,3)-
+
- Create a categorical variable with two levels (i.e.,
+
smallerandbigger)
+
+z = ifelse(x < y, 'smaller', 'bigger')-
+
- Create a categorical variable with three levels. An alternative
+approach would be to use the
Recodefunction described +below
+
-
+
- Convert an outlier to a missing value. For example, if we want to
+remove the maximum value from a variable called
salesthat +is equal to 400 we could use anifelsestatement and enter +the command below in theCreatebox. Press +returnandStoreto add the +sales_rcto the data. Note that if we had entered +saleson the left-hand side of the=sign the +original variable would have been overwritten
+
+sales_rc = ifelse(sales > 400, NA, sales)-
+
- If a respondent with ID 3 provided information on the wrong scale in
+a survey (e.g., income in $1s rather than in $1000s) we could use an
+
ifelsestatement and enter the command below in the +Createbox. As before, pressreturnand +Storeto addsales_rcto the data
+
+income_rc = ifelse(ID == 3, income/1000, income)-
+
- If multiple respondents made the same scaling mistake (e.g., those
+with ID 1, 3, and 15) we again use
Createand enter:
+
-
+
- If a date variable is in a format not available through the
+
Typemenu you can use theparse_date_time+function. For a date formatted as2-1-14you would specify +the command below (note that this format will also be parsed correctly +by themdyfunction in theTypemenu)
+
+date = parse_date_time(x, '%m%d%y')-
+
- Determine the time difference between two dates/times in +seconds +
+tdiff = as_duration(time2 - time1)-
+
- Extract the month from a date variable +
+m = month(date)-
+
- Other attributes that can be extracted from a date or date-time
+variable are
minute,hour,day, +week,quarter,year, +wday(for weekday). Forwdayand +monthit can be convenient to addlabel = TRUE+to the call. For example, to extract the weekday from a date variable +and use a label rather than a number
+
+wd = wday(date, label = TRUE)-
+
- Calculate the distance between two locations using lat-long +information +
+dist = as_distance(lat1, long1, lat2, long2)-
+
- Calculate quintiles for a variable
recencyby using the +xtilecommand. To create deciles replace5by +10.
+
+rec_iq = xtile(recency, 5)-
+
- To reverse the ordering of the quintiles created in 17 above use
+
rev = TRUE+
+
+rec_iq = xtile(recency, 5, rev = TRUE)-
+
- To remove text from entries in a character or factor variable use
+
subto remove only the first instance orgsub+to remove all instances. For example, suppose each row for a variable +bk_scorehas the letters “clv” before a number (e.g., +“clv150”). We could replace each occurrence of “clv” by “” as +follows:
+
+bk_score = sub("clv", "", bk_score)Note: For examples 7, 8, and 15 above you may need to change the new
+variable to type factor before using it for further
+analysis (see also Change type above)
Clean data +
+Remove missing values +
+Choose Remove missing from the
+Transformation type drop-down to eliminate rows with one or
+more missing values. Rows with missing values for
+Select variables will be removed. Press Store
+to change the data. If missing values were present you will see the
+number of observations in the data summary change (i.e., the value of
+n changes) as variables are selected.
Reorder or remove variables +
+Choose Reorder/Remove variables from the
+Transformation type drop-down. Drag-and-drop variables to
+reorder them in the data. To remove a variable click the \(\times\) symbol next to the label. Press
+Store to commit the changes.
Remove duplicates +
+It is common to have one or more variables in a dataset that have
+only unique values (i.e., no duplicates). Customer IDs, for example,
+should be unique unless the dataset contains multiple orders for the
+same customer. To remove duplicates select one or more variables to
+determine uniqueness. Choose Remove duplicates
+from the Transformation type drop-down and check how the
+displayed summary statistics change. Press Store to change
+the data. If there are duplicate rows you will see the number of
+observations in the data summary change (i.e., the value of n
+and n_distinct will change).
Show duplicates +
+If there are duplicates in the data use Show duplicates
+to get a better sense for the data points that have the same value in
+multiple rows. If you want to explore duplicates using the
+Data
+> View tab make sure to Store them in a
+different dataset (i.e., make sure not to overwrite the
+data you are working on). If you choose to show duplicates based on all
+columns in the data only one of the duplicate rows will be shown. These
+rows are exactly the same so showing 2 or 3 isn’t
+helpful. If, however, we are looking for duplicates based on a subset of
+the available variables Radiant will generate a dataset with
+all relevant rows.
Expand data +
+Expand grid +
+Create a dataset with all combinations of values for a selection of
+variables. This is useful to generate datasets for prediction in, for
+example,
+Model
+> Estimate > Linear regression (OLS) or
+Model
+> Estimate > Logistic regression (GLM). Suppose you want
+to create a dataset with all possible combinations of values for
+cut and color of a diamond. By selecting
+Expand grid from the Transformation type
+dropdown and cut and color in the
+Select variable(s) box we can see in the screenshot below
+that there are 35 possible combinations (i.e., cut has 5
+unique values and color has 7 unique values so 5 x 7
+combinations are possible). Choose a name for the new dataset (e.g.,
+diamonds_expand) and click the Store button to add it to
+the Datasets dropdown.
+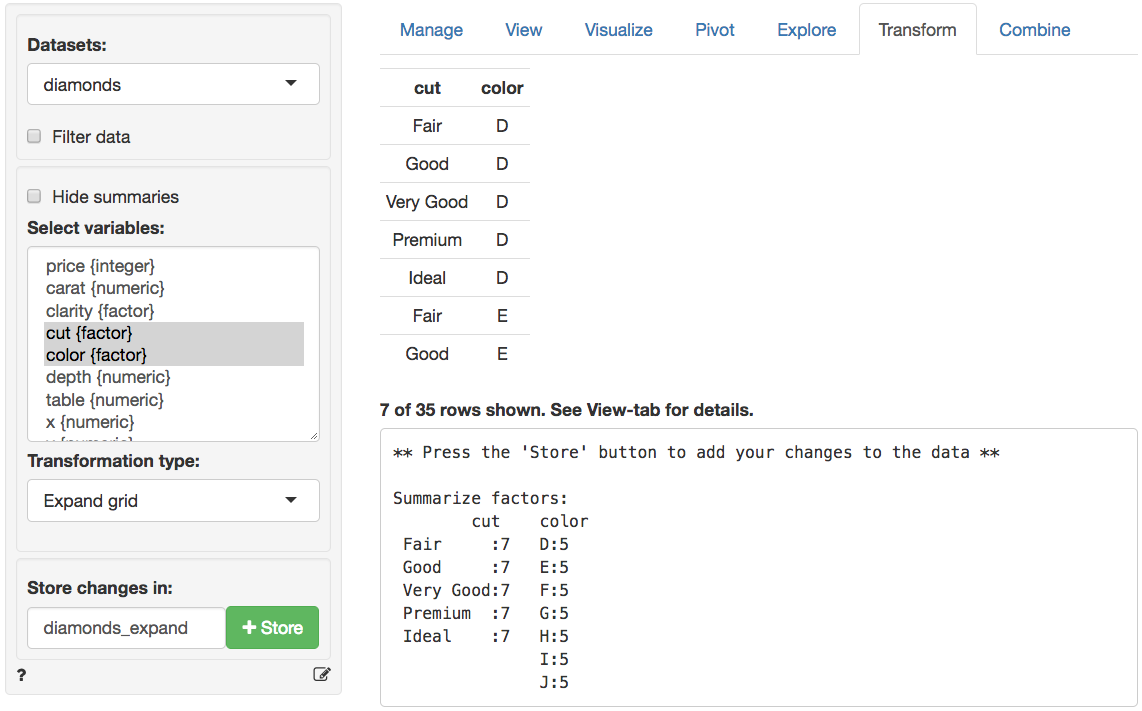
Split data +
+Holdout sample +
+To create a holdout sample based on a filter, select
+Holdout sample from the Transformation type
+dropdown. By default the opposite of the active filter is used.
+For example, if analysis is conducted on observations with
+date < '2014-12-13' then the holdout sample will contain
+rows with date >= '2014-12-13' if the
+Reverse filter box is checked.
Training variable +
+To create a variable that can be used to (randomly) filter a dataset
+for model training and testing, select Training variable
+from the Transformation type dropdown. Specify either the
+number of observations to use for training (e.g., set Size
+to 2000) or a proportion of observations to select (e.g., set
+Size to .7). The new variable will have a value
+1 for training and 0 test data.
It is also possible to select one or morel variables for
+blocking in random assignment to the training and test
+samples. This can help ensure that, for example, the proportion of
+positive and negative and negative cases (e.g., “buy” vs “no buy”) for a
+variable of interest is (almost) identical in the training and test
+sample.
Tidy data +
+Gather columns +
+Combine multiple variables into one column. If you have the
+diamonds dataset loaded, select cut and
+color in the Select variable(s) box after
+selecting Gather columns from the
+Transformation type dropdown. This will create new
+variables key and value. key has
+two levels (i.e., cut and color) and
+value captures all values in cut and
+color.
Spread column +
+Spread one column into multiple columns. The opposite of
+gather. For a detailed discussion about tidy data
+see the
+tidy-data
+vignette.
R-functions +
+For an overview of related R-functions used by Radiant to transform +data see +Data +> Transform
+ +View data in an interactive table (Data > +View)
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/view.Rmd
+ view.Rmd++Show data as an interactive table
+
Datasets +
+Choose one of the datasets from the Datasets dropdown.
+Files are loaded into Radiant through the Data > Manage
+tab.
Filter data +
+There are several ways to select a subset of the data to view. The
+Filter data box on the left (click the check-box) can be
+used with > and < symbols. You can also
+combine subset commands, for example, x > 3 & y == 2
+would show only those rows for which the variable x has
+values larger than 3 AND for which y is
+equal to 2. Note that in R, and most other programming languages,
+= is used to assign a value and == to
+determine if values are equal to each other. In contrast,
+!= is used to determine if two values are unequal.
+You can also use expressions that have an OR condition.
+For example, to select rows where Salary is smaller than
+$100,000 OR larger than $20,000 use
+Salary > 20000 | Salary < 100000. | is
+the symbol for OR and & is the symbol
+for AND
It is also possible to filter using dates. For example, to select
+rows with dates before June 1st, 2014 enter
+date < "2014-6-1" into the filter box and press
+return.
You can also use string matching to select rows. For example, type
+grepl('ood', cut) to select rows with Good or
+Very good cut. This search is case sensitive by default.
+For case insensitive search use
+grepl("GOOD", cut, ignore.case = TRUE). Type your statement
+in the Filter box and press return to see the result on
+screen or an error below the box if the expression is invalid.
It is important to note that these filters are persistent
+and will be applied to any analysis conducted through in Radiant. To
+deactivate a filter un-check the Filter data check-box. To
+remove a filter simply delete it.
| +Operator + | ++Description + | ++Example + | +
|---|---|---|
+<
+ |
++less than + | +
+price < 5000
+ |
+
+<=
+ |
++less than or equal to + | +
+carat <= 2
+ |
+
+>
+ |
++greater than + | +
+price > 1000
+ |
+
+>=
+ |
++greater than or equal to + | +
+carat >= 2
+ |
+
+==
+ |
++exactly equal to + | +
+cut == 'Fair'
+ |
+
+!=
+ |
++not equal to + | +
+cut != 'Fair'
+ |
+
+|
+ |
++x OR y + | +
+price > 10000 | cut == 'Premium'
+ |
+
+&
+ |
++x AND y + | +
+carat < 2 & cut == 'Fair'
+ |
+
+%in%
+ |
++x is one of y + | +
+cut %in% c('Fair', 'Good')
+ |
+
| +is.na + | ++is missing + | +
+is.na(price)
+ |
+
Filters can also be used with R-code to quickly view a sample from
+the selected dataset. For example, runif(n()) > .9 could
+be used to sample approximately 10% of the rows in the data and
+1:n() < 101 would select only the first 100 rows in the
+data.
Select variables to show +
+By default all columns in the data are shown. Click on any variable +to focus on it alone. To select several variables use the SHIFT and +ARROW keys on your keyboard. On a mac the CMD key can also be used to +select multiple variables. The same effect is achieved on windows using +the CTRL key. To select all variable use CTRL-A (or CMD-A on mac).
+Browse the data +
+By default only 10 rows of data are shown at a time. You can change
+this setting through the Show ... entries dropdown. Press
+the Next and Previous buttons at the
+bottom-right of the screen to page through the data.
Sort +
+Click on a column header in the table to sort the data. Clicking +again will toggle between sorting in ascending and descending order. To +sort on multiple columns at once press shift and then click on the 2nd, +3rd, etc. column to sort by.
+Column filters and Search +
+For variables that have a limited number of different values (i.e., a
+factor) you can select the levels to keep from the column filter below
+the variable name. For example, to filter on rows with ideal cut click
+in the box below the cut column header and select
+Ideal from the dropdown menu shown. You can also type a
+string into these column filters and then press return. Note that
+matching is case-insensitive. In fact, typing eal would
+produce the same result because the search will match any part of a
+string. Similarly, you can type a string to select rows based on
+character variables (e.g., street names).
For numeric variables the column filter boxes have some special
+features that make them almost as powerful as the
+Filter data box. For numeric and integer variables you can
+use ... to indicate a range. For example, to select
+price values between $500 and $2000 type
+500 ... 2000 and press return. The range is inclusive of
+the values typed. Furthermore, if we want to filter on
+carat 0.32 ... will show only diamonds with
+carat values larger than or equal to 0.32. Numeric variables also have a
+slider that you can use to define the range of values to keep.
If you want to get really fancy you can use the search box
+on the top right to search across all columns in the data using
+regular expressions. For example, to find all rows that
+have an entry in any column ending with the number 72 type
+72$ (i.e., the $ sign is used to indicate the
+end of an entry). For all rows with entries that start with 60 use
+^60 (i.e., the ^ is used to indicate the first
+character in an entry). Regular expressions are incredibly powerful for
+search but this is a big topic area. To learn more about
+regular expressions see this
+tutorial.
Store filters +
+It is important to note that column sorting, column filters, and
+search are not persistent. To store these settings for
+use in other parts of Radiant press the Store button. You
+can store the data and settings under a different dataset name by
+changing the value in the text input to the left of the
+Store button. This feature can also be used to select a
+subset of variables to keep. Just select the ones you want to keep and
+press the Store button. For more control over the variables
+you want to keep or remove and to specify their order in the dataset use
+the Data > Transform tab.
To download the data in csv format click the + icon on the top right of +your screen.
+Click the report ()
+icon on the bottom left of your screen or press ALT-enter
+on your keyboard to add the filter and sort commands used by Radiant to
+a (reproducible) report in
+Report
+> Rmd.
R-functions +
+For an overview of related R-functions used by Radiant to view, +search, and filter data see +Data +> View
+ +Visualize data (Data > Visualize)
+Vincent R. +Nijs, Rady School of Management (UCSD)
+ + + Source:vignettes/pkgdown/visualize.Rmd
+ visualize.Rmd++Visualize data
+
Filter data +
+Use the Filter data box to select (or omit) specific
+sets of rows from the data. See the help file for
+Data
+> View for details.
Plot-type +
+Select the plot type you want. For example, with the
+diamonds data loaded select Distribution and
+all (X) variables (use CTRL-a or CMD-a). This will create a histogram
+for all numeric variables and a bar-plot for all categorical variables
+in the data set. Density plots can only be used with numeric variables.
+Scatter plots are used to visualize the relationship between two
+variables. Select one or more variables to plot on the Y-axis and one or
+more variables to plot on the X-axis. If one of the variables is
+categorical (i.e., a {factor}) it should be specified as an X-variable.
+Information about additional variables can be added through the
+Color or Size dropdown. Line plots are similar
+to scatter plots but they connect-the-dots and are particularly useful
+for time-series data. Surface plots are similar to
+Heat maps and require 3 input variables: X, Y, and Fill.
+Bar plots are used to show the relationship between a categorical (or
+integer) variable (X) and the (mean) value of a numeric variable (Y).
+Box-plots are also used when we have a numeric Y-variable and a
+categorical X-variable. They are more informative than bar charts but
+also require a bit more effort to evaluate.
++Note that when a categorical variable (
+factor) is +selected as theY-variablein a Bar chart it will be +converted to a numeric variable if required for the selected function. +If the factor levels are numeric these will be used in all calculations. +Since the mean, standard deviation, etc. are not relevant for non-binary +categorical variables, these will be converted to 0-1 (binary) variables +where the first level is coded as 1 and all other levels as 0. For +example, if we selectcolorfrom thediamonds+data as the Y-variable, andmeanas the function to apply, +then each bar will represent the proportion of observations with the +valueD.
Box plots +
+The upper and lower “hinges” of the box correspond to the first and +third quartiles (the 25th and 75th percentiles) in the data. The middle +hinge is the median value of the data. The upper whisker extends from +the upper hinge (i.e., the top of the box) to the highest value in the +data that is within 1.5 x IQR of the upper hinge. IQR is the +inter-quartile range, or distance, between the 25th and 75th percentile. +The lower whisker extends from the lower hinge to the lowest value in +the data within 1.5 x IQR of the lower hinge. Data beyond the end of the +whiskers could be outliers and are plotted as points (as suggested by +Tukey).
+In sum: 1. The lower whisker extends from Q1 to max(min(data), Q1 - +1.5 x IQR) 2. The upper whisker extends from Q3 to min(max(data), Q3 + +1.5 x IQR)
+where Q1 is the 25th percentile and Q3 is the 75th percentile. You +may have to read the two bullets above a few times before it sinks in. +The plot below should help to explain the structure of the box plot.
+
+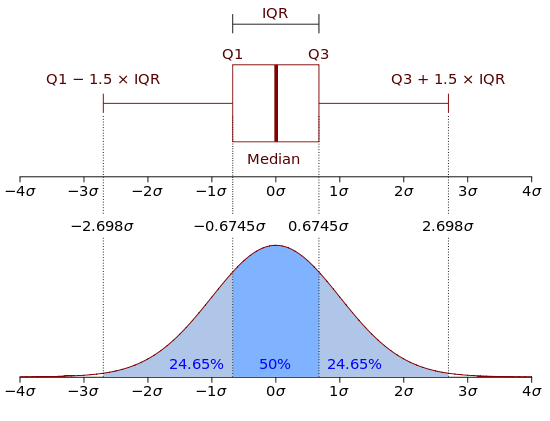
Sub-plots and heat-maps +
+Facet row and Facet column can be used to
+split the data into different groups and create separate plots for each
+group.
If you select a scatter or line plot a Color drop-down
+will be shown. Selecting a Color variable will create a
+type of heat-map where the colors are linked to the values of the
+Color variable. Selecting a categorical variable from the
+Color dropdown for a line plot will split the data into
+groups and will show a line of a different color for each group.
Line, loess, and jitter +
+To add a linear or non-linear regression line to a scatter plot check
+the Line and/or Loess boxes. If your data take
+on a limited number of values, Jitter can be useful to get
+a better feel for where most of the data points are located.
+Jitter-ing simply adds a small random value to each data
+point so they do not overlap completely in the plot(s).
Axis scale +
+The relationship between variables depicted in a scatter plot may be
+non-linear. There are numerous transformations we might apply to the
+data so this relationship becomes (approximately) linear (see
+Data
+> Transform) and easier to estimate using, for example,
+Model
+> Estimate > Linear regression (OLS). Perhaps the most
+common data transformation applied to business data is the (natural)
+logarithm. To see if log transformation(s) may be appropriate for your
+data check the Log X and/or Log Y boxes (e.g.,
+for a scatter or bar plot).
By default the scale of the Y-axis is the same across sub-plots when
+using Facet row. To allow the Y-axis to be specific to each
+sub-plot click the Scale-y check-box.
Plot height and width +
+To make plots bigger or smaller adjust the values in the height and +width boxes on the bottom left of the screen.
+Keep plots +
+The best way to keep/store plots is to generate a
+visualize command by clicking the report
+() icon on the bottom
+left of your screen or by pressing ALT-enter on your
+keyboard. Alternatively, click the
+ icon on the top right of
+your screen to save a png-file to disk.
Customizing plots in Report > Rmd +
+To customize a plot first generate the visualize command
+by clicking the report
+() icon on the bottom
+left of your screen or by pressing ALT-enter on your
+keyboard. The example below illustrates how to customize a command in
+the
+Report
+> Rmd tab. Notice that custom is set to
+TRUE.
+visualize(diamonds, yvar = "price", xvar = "carat", type = "scatter", custom = TRUE) +
+ labs(
+ title = "A scatterplot",
+ y = "Price in $",
+ x = "Carats"
+ )The default resolution for plots is 144 dots per inch (dpi). You can +change this setting up or down in Report > Rmd. For example, +the code-chunk header below ensures the plot will be 7” wide, 3.5” tall, +with a resolution of 600 dpi.
+```{r fig.width = 7, fig.height = 3.5, dpi = 600}
If you have the svglite package installed, the
+code-chunk header below will produce graphs in high quality
+svg format.
```{r fig.width = 7, fig.height = 3.5, dev = "svglite"}
Some common customization commands:
+-
+
- Add a title:
+ labs(title = "my title")+
+ - Add a sub-title:
+ labs(subtitle = "my sub-title")+
+ - Add a caption below figure:
+
+ labs(caption = "Based on data from ...")+
+ - Change label:
+ labs(x = "my X-axis label")or ++ labs(y = "my Y-axis label")+
+ - Remove all legends:
+
+ theme(legend.position = "none")+
+ - Change legend title:
+ labs(color = "New legend title")+or+ labs(fill = "New legend title")+
+ - Rotate tick labels:
+
+ theme(axis.text.x = element_text(angle = 90, hjust = 1))+
+ - Set plot limits:
+ ylim(5000, 8000)or ++ xlim("VS1","VS2")+
+ - Remove size legend:
+ scale_size(guide = "none")+
+ - Change size range:
+ scale_size(range=c(1,6))+
+ - Draw a horizontal line:
+
+ geom_hline(yintercept = 0.1)+
+ - Draw a vertical line:
+ geom_vline(xintercept = 8)+
+ - Scale the y-axis as a percentage:
+
+ scale_y_continuous(labels = scales::percent)+
+ - Scale the y-axis in millions:
+
+ scale_y_continuous(labels = scales::unit_format(unit = "M", scale = 1e-6))+
+ - Display y-axis in $’s:
+
+ scale_y_continuous(labels = scales::dollar_format())+
+ - Use
,as a thousand separator for the y-axis: ++ scale_y_continuous(labels = scales::comma)+
+
For more on how to customize plots for communication see +http://r4ds.had.co.nz/graphics-for-communication.html.
+See also the ggplot2 documentation site +https://ggplot2.tidyverse.org.
+Suppose we create a set of three bar charts in Data >
+Visualize using the Diamond data. To add a title above
+the group of plots and impose a one-column layout we could use
+patchwork as follows:
+plot_list <- visualize(
+ diamonds,
+ xvar = c("clarity", "cut", "color"),
+ yvar = "price",
+ type = "bar",
+ custom = TRUE
+)
+wrap_plots(plot_list, ncol = 1) + plot_annotation(title = "Three bar plots")See the patchwork +documentation site for additional information on how to customize +groups of plots.
+Making plots interactive in Report > Rmd +
+It is possible to transform (most) plots generated in Radiant into
+interactive graphics using the plotly library. After
+setting custom = TRUE you can use the ggplotly
+function to convert a single plot. See example below:
+visualize(diamonds, xvar = c("price", "carat", "clarity", "cut"), custom = TRUE) %>%
+ ggplotly() %>%
+ render()If more than one plot is created, you can use the
+subplot function from the plotly package.
+Provide a value for the nrows argument to setup the plot
+layout grid. In the example below four plots are created. Because
+nrow = 2 the plots will be displayed in a 2 X 2 grid.
+visualize(diamonds, xvar = c("carat", "clarity", "cut", "color"), custom = TRUE) %>%
+ subplot(nrows = 2) %>%
+ render()For additional information on the plotly library see the
+links below:
-
+
- Getting started: +https://plot.ly/r/getting-started/ + +
- Reference: +https://plot.ly/r/reference/ + +
- Book: +https://cpsievert.github.io/plotly_book + +
- Code: +https://github.com/ropensci/plotly + +
R-functions +
+For an overview of related R-functions used by Radiant to visualize +data see +Data +> Visualize
+ +`, then ``, etc.) that has more than one element. Defaults to 1 (for ``).
+ getTopLevel: function($scope) {
+ for (var i = 1; i <= 6; i++) {
+ var $headings = this.findOrFilter($scope, 'h' + i);
+ if ($headings.length > 1) {
+ return i;
+ }
+ }
+
+ return 1;
+ },
+
+ // returns the elements for the top level, and the next below it
+ getHeadings: function($scope, topLevel) {
+ var topSelector = 'h' + topLevel;
+
+ var secondaryLevel = topLevel + 1;
+ var secondarySelector = 'h' + secondaryLevel;
+
+ return this.findOrFilter($scope, topSelector + ',' + secondarySelector);
+ },
+
+ getNavLevel: function(el) {
+ return parseInt(el.tagName.charAt(1), 10);
+ },
+
+ populateNav: function($topContext, topLevel, $headings) {
+ var $context = $topContext;
+ var $prevNav;
+
+ var helpers = this;
+ $headings.each(function(i, el) {
+ var $newNav = helpers.generateNavItem(el);
+ var navLevel = helpers.getNavLevel(el);
+
+ // determine the proper $context
+ if (navLevel === topLevel) {
+ // use top level
+ $context = $topContext;
+ } else if ($prevNav && $context === $topContext) {
+ // create a new level of the tree and switch to it
+ $context = helpers.createChildNavList($prevNav);
+ } // else use the current $context
+
+ $context.append($newNav);
+
+ $prevNav = $newNav;
+ });
+ },
+
+ parseOps: function(arg) {
+ var opts;
+ if (arg.jquery) {
+ opts = {
+ $nav: arg
+ };
+ } else {
+ opts = arg;
+ }
+ opts.$scope = opts.$scope || $(document.body);
+ return opts;
+ }
+ },
+
+ // accepts a jQuery object, or an options object
+ init: function(opts) {
+ opts = this.helpers.parseOps(opts);
+
+ // ensure that the data attribute is in place for styling
+ opts.$nav.attr('data-toggle', 'toc');
+
+ var $topContext = this.helpers.createChildNavList(opts.$nav);
+ var topLevel = this.helpers.getTopLevel(opts.$scope);
+ var $headings = this.helpers.getHeadings(opts.$scope, topLevel);
+ this.helpers.populateNav($topContext, topLevel, $headings);
+ }
+ };
+
+ $(function() {
+ $('nav[data-toggle="toc"]').each(function(i, el) {
+ var $nav = $(el);
+ Toc.init($nav);
+ });
+ });
+})();
diff --git a/radiant.data/docs/docsearch.css b/radiant.data/docs/docsearch.css
new file mode 100644
index 0000000000000000000000000000000000000000..e5f1fe1dfa2c34c51fe941829b511acd8c763301
--- /dev/null
+++ b/radiant.data/docs/docsearch.css
@@ -0,0 +1,148 @@
+/* Docsearch -------------------------------------------------------------- */
+/*
+ Source: https://github.com/algolia/docsearch/
+ License: MIT
+*/
+
+.algolia-autocomplete {
+ display: block;
+ -webkit-box-flex: 1;
+ -ms-flex: 1;
+ flex: 1
+}
+
+.algolia-autocomplete .ds-dropdown-menu {
+ width: 100%;
+ min-width: none;
+ max-width: none;
+ padding: .75rem 0;
+ background-color: #fff;
+ background-clip: padding-box;
+ border: 1px solid rgba(0, 0, 0, .1);
+ box-shadow: 0 .5rem 1rem rgba(0, 0, 0, .175);
+}
+
+@media (min-width:768px) {
+ .algolia-autocomplete .ds-dropdown-menu {
+ width: 175%
+ }
+}
+
+.algolia-autocomplete .ds-dropdown-menu::before {
+ display: none
+}
+
+.algolia-autocomplete .ds-dropdown-menu [class^=ds-dataset-] {
+ padding: 0;
+ background-color: rgb(255,255,255);
+ border: 0;
+ max-height: 80vh;
+}
+
+.algolia-autocomplete .ds-dropdown-menu .ds-suggestions {
+ margin-top: 0
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion {
+ padding: 0;
+ overflow: visible
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--category-header {
+ padding: .125rem 1rem;
+ margin-top: 0;
+ font-size: 1.3em;
+ font-weight: 500;
+ color: #00008B;
+ border-bottom: 0
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--wrapper {
+ float: none;
+ padding-top: 0
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--subcategory-column {
+ float: none;
+ width: auto;
+ padding: 0;
+ text-align: left
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--content {
+ float: none;
+ width: auto;
+ padding: 0
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--content::before {
+ display: none
+}
+
+.algolia-autocomplete .ds-suggestion:not(:first-child) .algolia-docsearch-suggestion--category-header {
+ padding-top: .75rem;
+ margin-top: .75rem;
+ border-top: 1px solid rgba(0, 0, 0, .1)
+}
+
+.algolia-autocomplete .ds-suggestion .algolia-docsearch-suggestion--subcategory-column {
+ display: block;
+ padding: .1rem 1rem;
+ margin-bottom: 0.1;
+ font-size: 1.0em;
+ font-weight: 400
+ /* display: none */
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--title {
+ display: block;
+ padding: .25rem 1rem;
+ margin-bottom: 0;
+ font-size: 0.9em;
+ font-weight: 400
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--text {
+ padding: 0 1rem .5rem;
+ margin-top: -.25rem;
+ font-size: 0.8em;
+ font-weight: 400;
+ line-height: 1.25
+}
+
+.algolia-autocomplete .algolia-docsearch-footer {
+ width: 110px;
+ height: 20px;
+ z-index: 3;
+ margin-top: 10.66667px;
+ float: right;
+ font-size: 0;
+ line-height: 0;
+}
+
+.algolia-autocomplete .algolia-docsearch-footer--logo {
+ background-image: url("data:image/svg+xml;utf8,");
+ background-repeat: no-repeat;
+ background-position: 50%;
+ background-size: 100%;
+ overflow: hidden;
+ text-indent: -9000px;
+ width: 100%;
+ height: 100%;
+ display: block;
+ transform: translate(-8px);
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--highlight {
+ color: #FF8C00;
+ background: rgba(232, 189, 54, 0.1)
+}
+
+
+.algolia-autocomplete .algolia-docsearch-suggestion--text .algolia-docsearch-suggestion--highlight {
+ box-shadow: inset 0 -2px 0 0 rgba(105, 105, 105, .5)
+}
+
+.algolia-autocomplete .ds-suggestion.ds-cursor .algolia-docsearch-suggestion--content {
+ background-color: rgba(192, 192, 192, .15)
+}
diff --git a/radiant.data/docs/docsearch.js b/radiant.data/docs/docsearch.js
new file mode 100644
index 0000000000000000000000000000000000000000..b35504cd3a282816130a16881f3ebeead9c1bcb4
--- /dev/null
+++ b/radiant.data/docs/docsearch.js
@@ -0,0 +1,85 @@
+$(function() {
+
+ // register a handler to move the focus to the search bar
+ // upon pressing shift + "/" (i.e. "?")
+ $(document).on('keydown', function(e) {
+ if (e.shiftKey && e.keyCode == 191) {
+ e.preventDefault();
+ $("#search-input").focus();
+ }
+ });
+
+ $(document).ready(function() {
+ // do keyword highlighting
+ /* modified from https://jsfiddle.net/julmot/bL6bb5oo/ */
+ var mark = function() {
+
+ var referrer = document.URL ;
+ var paramKey = "q" ;
+
+ if (referrer.indexOf("?") !== -1) {
+ var qs = referrer.substr(referrer.indexOf('?') + 1);
+ var qs_noanchor = qs.split('#')[0];
+ var qsa = qs_noanchor.split('&');
+ var keyword = "";
+
+ for (var i = 0; i < qsa.length; i++) {
+ var currentParam = qsa[i].split('=');
+
+ if (currentParam.length !== 2) {
+ continue;
+ }
+
+ if (currentParam[0] == paramKey) {
+ keyword = decodeURIComponent(currentParam[1].replace(/\+/g, "%20"));
+ }
+ }
+
+ if (keyword !== "") {
+ $(".contents").unmark({
+ done: function() {
+ $(".contents").mark(keyword);
+ }
+ });
+ }
+ }
+ };
+
+ mark();
+ });
+});
+
+/* Search term highlighting ------------------------------*/
+
+function matchedWords(hit) {
+ var words = [];
+
+ var hierarchy = hit._highlightResult.hierarchy;
+ // loop to fetch from lvl0, lvl1, etc.
+ for (var idx in hierarchy) {
+ words = words.concat(hierarchy[idx].matchedWords);
+ }
+
+ var content = hit._highlightResult.content;
+ if (content) {
+ words = words.concat(content.matchedWords);
+ }
+
+ // return unique words
+ var words_uniq = [...new Set(words)];
+ return words_uniq;
+}
+
+function updateHitURL(hit) {
+
+ var words = matchedWords(hit);
+ var url = "";
+
+ if (hit.anchor) {
+ url = hit.url_without_anchor + '?q=' + escape(words.join(" ")) + '#' + hit.anchor;
+ } else {
+ url = hit.url + '?q=' + escape(words.join(" "));
+ }
+
+ return url;
+}
diff --git a/radiant.data/docs/docsearch.json b/radiant.data/docs/docsearch.json
new file mode 100644
index 0000000000000000000000000000000000000000..715937648ea49c8f2e3f10d27fa5c14b75d844a8
--- /dev/null
+++ b/radiant.data/docs/docsearch.json
@@ -0,0 +1,95 @@
+{
+ "index_name": "radiant_data",
+ "start_urls": [
+ {
+ "url": "https://radiant-rstats.github.io/radiant.data/index.html",
+ "selectors_key": "homepage",
+ "tags": [
+ "homepage"
+ ]
+ },
+ {
+ "url": "https://radiant-rstats.github.io/radiant.data/reference",
+ "selectors_key": "reference",
+ "tags": [
+ "reference"
+ ]
+ },
+ {
+ "url": "https://radiant-rstats.github.io/radiant.data/articles",
+ "selectors_key": "articles",
+ "tags": [
+ "articles"
+ ]
+ }
+ ],
+ "stop_urls": [
+ "/reference/$",
+ "/reference/index.html",
+ "/articles/$",
+ "/articles/index.html"
+ ],
+ "sitemap_urls": [
+ "https://radiant-rstats.github.io/radiant.data/sitemap.xml"
+ ],
+ "selectors": {
+ "homepage": {
+ "lvl0": {
+ "selector": ".contents h1",
+ "default_value": "radiant.data Home page"
+ },
+ "lvl1": {
+ "selector": ".contents h2"
+ },
+ "lvl2": {
+ "selector": ".contents h3",
+ "default_value": "Context"
+ },
+ "lvl3": ".ref-arguments td, .ref-description",
+ "text": ".contents p, .contents li, .contents .pre"
+ },
+ "reference": {
+ "lvl0": {
+ "selector": ".contents h1"
+ },
+ "lvl1": {
+ "selector": ".contents .name",
+ "default_value": "Argument"
+ },
+ "lvl2": {
+ "selector": ".ref-arguments th",
+ "default_value": "Description"
+ },
+ "lvl3": ".ref-arguments td, .ref-description",
+ "text": ".contents p, .contents li"
+ },
+ "articles": {
+ "lvl0": {
+ "selector": ".contents h1"
+ },
+ "lvl1": {
+ "selector": ".contents .name"
+ },
+ "lvl2": {
+ "selector": ".contents h2, .contents h3",
+ "default_value": "Context"
+ },
+ "text": ".contents p, .contents li"
+ }
+ },
+ "selectors_exclude": [
+ ".dont-index"
+ ],
+ "min_indexed_level": 2,
+ "custom_settings": {
+ "separatorsToIndex": "_",
+ "attributesToRetrieve": [
+ "hierarchy",
+ "content",
+ "anchor",
+ "url",
+ "url_without_anchor"
+ ]
+ }
+}
+
diff --git a/radiant.data/docs/index.html b/radiant.data/docs/index.html
new file mode 100644
index 0000000000000000000000000000000000000000..3c25a5ddaedeb3ee2c75f8e391faa7ade42fa79d
--- /dev/null
+++ b/radiant.data/docs/index.html
@@ -0,0 +1,365 @@
+
+
+
+
+
+
+
+Data Menu for Radiant: Business Analytics using R and Shiny • radiant.data
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+Radiant is an open-source platform-independent browser-based interface for business analytics in R. The application is based on the Shiny package and can be run locally or on a server. Radiant was developed by Vincent Nijs. Please use the issue tracker on GitHub to suggest enhancements or report problems: https://github.com/radiant-rstats/radiant.data/issues. For other questions and comments please use radiant@rady.ucsd.edu.
+
+Key features
+
+
+- Explore: Quickly and easily summarize, visualize, and analyze your data
+- Cross-platform: It runs in a browser on Windows, Mac, and Linux
+- Reproducible: Recreate results and share work with others as a state file or an Rmarkdown report
+- Programming: Integrate Radiant’s analysis functions with your own R-code
+- Context: Data and examples focus on business applications
+
+
+
+Playlists
+
+There are two youtube playlists with video tutorials. The first provides a general introduction to key features in Radiant. The second covers topics relevant in a course on business analytics (i.e., Probability, Decision Analysis, Hypothesis Testing, Linear Regression, and Simulation).
+
+
+
+Explore
+
+Radiant is interactive. Results update immediately when inputs are changed (i.e., no separate dialog boxes) and/or when a button is pressed (e.g., Estimate in Model > Estimate > Logistic regression (GLM)). This facilitates rapid exploration and understanding of the data.
+
+
+Cross-platform
+
+Radiant works on Windows, Mac, or Linux. It can run without an Internet connection and no data will leave your computer. You can also run the app as a web application on a server.
+
+
+Reproducible
+
+To conduct high-quality analysis, simply saving output is not enough. You need the ability to reproduce results for the same data and/or when new data become available. Moreover, others may want to review your analysis and results. Save and load the state of the application to continue your work at a later time or on another computer. Share state files with others and create reproducible reports using Rmarkdown. See also the section on Saving and loading state below
+If you are using Radiant on a server you can even share the URL (include the SSUID) with others so they can see what you are working on. Thanks for this feature go to Joe Cheng.
+
+
+Programming
+
+Although Radiant’s web-interface can handle quite a few data and analysis tasks, you may prefer to write your own R-code. Radiant provides a bridge to programming in R(studio) by exporting the functions used for analysis (i.e., you can conduct your analysis using the Radiant web-interface or by calling Radiant’s functions directly from R-code). For more information about programming with Radiant see the programming page on the documentation site.
+
+
+
+
+How to install Radiant
+
+
+In Rstudio you can start and update Radiant through the Addins menu at the top of the screen. To install the latest version of Radiant for Windows or Mac, with complete documentation for off-line access, open R(studio) and copy-and-paste the command below:
+
+options(repos = c(RSM = "https://radiant-rstats.github.io/minicran", CRAN = "https://cloud.r-project.org"))
+install.packages("radiant")
+Once all packages are installed, select Start radiant from the Addins menu in Rstudio or use the command below to launch the app:
+
+radiant::radiant()
+To launch Radiant in Rstudio’s viewer pane use the command below:
+
+radiant::radiant_viewer()
+To launch Radiant in an Rstudio Window use the command below:
+
+radiant::radiant_window()
+To easily update Radiant and the required packages, install the radiant.update package using:
+
+options(repos = c(RSM = "https://radiant-rstats.github.io/minicran", CRAN = "https://cloud.r-project.org"))
+install.packages("remotes")
+remotes::install_github("radiant-rstats/radiant.update", upgrade = "never")
+Then select Update radiant from the Addins menu in Rstudio or use the command below:
+
+radiant.update::radiant.update()
+See the installing radiant page additional for details.
+Optional: You can also create a launcher on your Desktop to start Radiant by typing radiant::launcher() in the R(studio) console and pressing return. A file called radiant.bat (windows) or radiant.command (mac) will be created that you can double-click to start Radiant in your default browser. The launcher command will also create a file called update_radiant.bat (windows) or update_radiant.command (mac) that you can double-click to update Radiant to the latest release.
+When Radiant starts you will see data on diamond prices. To close the application click the icon in the navigation bar and then click Stop. The Radiant process will stop and the browser window will close (Chrome) or gray-out.
+
+
+Documentation
+
+Documentation and tutorials are available at https://radiant-rstats.github.io/docs/ and in the Radiant web interface (the icons on each page and the icon in the navigation bar).
+Individual Radiant packages also each have their own pkgdown sites:
+
+- http://radiant-rstats.github.io/radiant
+- http://radiant-rstats.github.io/radiant.data
+- http://radiant-rstats.github.io/radiant.design
+- http://radiant-rstats.github.io/radiant.basics
+- http://radiant-rstats.github.io/radiant.model
+- http://radiant-rstats.github.io/radiant.multivariate
+
+Want some help getting started? Watch the tutorials on the documentation site.
+
+
+Reporting issues
+
+Please use the GitHub issue tracker at github.com/radiant-rstats/radiant/issues if you have any problems using Radiant.
+
+
+Try Radiant online
+
+Not ready to install Radiant on your computer? Try it online at the link below:
+https://vnijs.shinyapps.io/radiant
+Do not upload sensitive data to this public server. The size of data upload has been restricted to 10MB for security reasons.
+
+
+Running Radiant on shinyapps.io
+
+To run your own instance of Radiant on shinyapps.io first install Radiant and its dependencies. Then clone the radiant repo and ensure you have the latest version of the Radiant packages installed by running radiant/inst/app/for.shinyapps.io.R. Finally, open radiant/inst/app/ui.R and deploy the application.
+
+
+Running Radiant on shiny-server
+
+You can also host Radiant using shiny-server. First, install radiant on the server using the command below:
+
+options(repos = c(RSM = "https://radiant-rstats.github.io/minicran", CRAN = "https://cloud.r-project.org"))
+install.packages("radiant")
+Then clone the radiant repo and point shiny-server to the inst/app/ directory. As a courtesy, please let me know if you intend to use Radiant on a server.
+When running Radiant on a server, by default, file uploads are limited to 10MB and R-code in Report > Rmd and Report > R will not be evaluated for security reasons. If you have sudo access to the server and have appropriate security in place you can change these settings by adding the following lines to .Rprofile for the shiny user on the server.
+
+
+
+Running Radiant in the cloud (e.g., AWS)
+
+To run radiant in the cloud you can use the customized Docker container. See https://github.com/radiant-rstats/docker for details
+
+
+Saving and loading state
+
+To save your analyses save the state of the app to a file by clicking on the icon in the navbar and then on Save radiant state file (see also the Data > Manage tab). You can open this state file at a later time or on another computer to continue where you left off. You can also share the file with others that may want to replicate your analyses. As an example, load the state file radiant-example.state.rda by clicking on the icon in the navbar and then on Load radiant state file. Go to Data > View and Data > Visualize to see some of the settings from the previous “state” of the app. There is also a report in Report > Rmd that was created using the Radiant interface. The html file radiant-example.nb.html contains the output.
+A related feature in Radiant is that state is maintained if you accidentally navigate to another web page, close (and reopen) the browser, and/or hit refresh. Use Refresh in the menu in the navigation bar to return to a clean/new state.
+Loading and saving state also works with Rstudio. If you start Radiant from Rstudio and use > Stop to stop the app, lists called r_data, r_info, and r_state will be put into Rstudio’s global workspace. If you start radiant again using radiant::radiant() it will use these lists to restore state. Also, if you load a state file directly into Rstudio it will be used when you start Radiant to recreate a previous state.
+Technical note: Loading state works as follows in Radiant: When an input is initialized in a Shiny app you set a default value in the call to, for example, numericInput. In Radiant, when a state file has been loaded and an input is initialized it looks to see if there is a value for an input of that name in a list called r_state. If there is, this value is used. The r_state list is created when saving state using reactiveValuesToList(input). An example of a call to numericInput is given below where the state_init function from radiant.R is used to check if a value from r_state can be used.
+
+numericInput("sm_comp_value", "Comparison value:", state_init("sm_comp_value", 0))
+
+
+Source code
+
+The source code for the radiant application is available on GitHub at https://github.com/radiant-rstats. radiant.data, offers tools to load, save, view, visualize, summarize, combine, and transform data. radiant.design builds on radiant.data and adds tools for experimental design, sampling, and sample size calculation. radiant.basics covers the basics of statistical analysis (e.g., comparing means and proportions, cross-tabs, correlation, etc.) and includes a probability calculator. radiant.model covers model estimation (e.g., logistic regression and neural networks), model evaluation (e.g., gains chart, profit curve, confusion matrix, etc.), and decision tools (e.g., decision analysis and simulation). Finally, radiant.multivariate includes tools to generate brand maps and conduct cluster, factor, and conjoint analysis.
+These tools are used in the Business Analytics, Quantitative Analysis, Research for Marketing Decisions, Applied Market Research, Consumer Behavior, Experiments in Firms, Pricing, Pricing Analytics, and Customer Analytics classes at the Rady School of Management (UCSD).
+
+
+Credits
+
+Radiant would not be possible without R and Shiny. I would like to thank Joe Cheng, Winston Chang, and Yihui Xie for answering questions, providing suggestions, and creating amazing tools for the R community. Other key components used in Radiant are ggplot2, dplyr, tidyr, magrittr, broom, shinyAce, shinyFiles, rmarkdown, and DT. For an overview of other packages that Radiant relies on please see the about page.
+
+
+License
+
+Radiant is licensed under the AGPLv3. As a summary, the AGPLv3 license requires, attribution, including copyright and license information in copies of the software, stating changes if the code is modified, and disclosure of all source code. Details are in the COPYING file.
+The documentation, images, and videos for the radiant.data package are licensed under the creative commons attribution and share-alike license CC-BY-SA. All other documentation and videos on this site, as well as the help files for radiant.design, radiant.basics, radiant.model, and radiant.multivariate, are licensed under the creative commons attribution, non-commercial, share-alike license CC-NC-SA.
+If you are interested in using any of the radiant packages please email me at radiant@rady.ucsd.edu
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/radiant.data/docs/link.svg b/radiant.data/docs/link.svg
new file mode 100644
index 0000000000000000000000000000000000000000..88ad82769b87f10725c57dca6fcf41b4bffe462c
--- /dev/null
+++ b/radiant.data/docs/link.svg
@@ -0,0 +1,12 @@
+
+
+
diff --git a/radiant.data/docs/news/index.html b/radiant.data/docs/news/index.html
new file mode 100644
index 0000000000000000000000000000000000000000..82cfa04db83273ec8c0e4369d9df25cb8517e3c8
--- /dev/null
+++ b/radiant.data/docs/news/index.html
@@ -0,0 +1,727 @@
+
+Changelog • radiant.data
+
+
+
+
+
+
+
+
+
+
+ Changelog
+ Source: NEWS.md
+
+
+
+
+radiant.data 1.5.12023-01-09
+- Added features in the UI to facilitate persistent filters for filtered, sorted, and sliced data
+- Improvements to screenshot feature:
+
- Navigation bar is omitted and the image is adjusted to the length of the UI.
+- html2canvas.js is now included so users can take screenshot when offline
+
+- Added a convenience function
add_description to add a description attribute to a data.frame in markdown format
+- Line graphs treated more similarly to bar-graphs:
+
- Can have a binary factor variable on the y-axis
+- Y-variable only line are now also possible
+
+- Removed all references to
aes_string which is being deprecated in ggplot soon
+- Improved cleanup after Radiant UI is closed
+
+
+
+radiant.data 1.4.6
+- gsub(“[-]”, ““, text) is no longer valid in R 4.2.0 and above. Non-asci symbols will now be escaped using stringi::stri_trans_general when needed
+
+
+radiant.data 1.4.52022-09-08
+- Add scrolling for dropdown menus that might extend past the edge of the screen
+- Addressed warning messages about Font Awesome icons not existing
+- gsub(“[-]”, ““, text) is no longer valid in R 4.2.0 and above. Non-asci symbols will now be escaped using stringi when needed
+
+
+radiant.data 1.4.42022-07-19
+- Added option to create screenshots of settings on a page. Approach is inspired by the snapper package by @yonicd
+- Added contact request for users on Radiant startup
+- Fix issue with R_ZIPCMD when 7zip is on the path but not being recognized by R
+
+
+radiant.data 1.4.22022-05-25
+- Use
all for is.null and is.na if object length can be greater than 1 as required in R 4.2.0
+
+
+radiant.data 1.4.12021-11-12
+- Setup to allow formatting of the shiny interface with bootstrap 4
+- Addressing
is_empty function clash with rlang
+
+- Upgrading
shiny dependency to 1.6.0 and fixing project text alignment issue (@cpsievert, https://github.com/radiant-rstats/radiant.data/pull/28)
+
+
+radiant.data 1.3.122020-11-27
+- Fixes related to breaking changes in
magrittr
+
+- Fixes related to changes in
readr argument names
+- Fix to launch radiant in a “windows”
+
+
+radiant.data 1.3.102020-08-07
+- Add Google Drive to the default set of directories to explore if available
+- Add back functionality to convert a column to type
ts in Data > Transform now that this is again supported by dplyr 1.0.1
+
+
+radiant.data 1.3.92020-06-16
+- Fix for using the
date function from the lubridate package in a filter
+- Removed functionality to convert a column to type
ts as this is not supported by dplyr 1.0.0 and vctrs 0.3.1
+- Updated documentation using https://github.com/r-lib/roxygen2/pull/1109
+
+
+
+radiant.data 1.3.6
+- Updated styling for formatting for modals (e.g., help pages) that will also allow improved sizing of the (shinyFiles) file browser
+- Fix for
\r line-endings in Report > Rmd on Windows. Issue was most likely to occur when copy-and-pasting text from PDF into Report > Rmd.
+
+
+
+radiant.data 1.3.3
+- Function to calculate “mode”
+- Fix for “spread” in Data > Transform with column name includes “.”
+
+
+radiant.data 1.3.1
+- If radiant is not opened from an Rstudio project, use the working directory at launch as the base directory for the application
+
+
+radiant.data 1.3.0
+- Updated styling of Notebook and HTML reports (cosmo + zenburn)
+- Documentation updates to link to new video tutorials
+- Use
patchwork for grouping multiple plots together
+- Apply
refactor to any type in the Data > Transform UI
+- Fix for
weighted.sd when missing values differ for x and weights
+- Avoid resetting the “Column header” to its default value in Data > Explore when other settings are changed.
+
+
+radiant.data 1.2.3
+- Fix for Data > Transform > Spread when no variables are selected
+- Set
debounce to 0 for all shinyAce editors
+
+
+radiant.data 1.2.2
+- Use
zenburn for code highlighting in Notebook and HTML report from Report > Rmd
+
+- Clean up “sf_volumes” from the when radiant is stopped
+
+
+radiant.data 1.2.0
+- Update action buttons that initiate a calculation when one or more relevant inputs are changed. For example, when a model should be re-estimated because the set of explanatory variables was changed by the user, a spinning “refresh” icon will be shown
+
+
+radiant.data 1.1.8
+- Changed default
quantile algorithm used in the xtile function from number 2 to 7. See the help for stats::quantile for details
+- Added
me and meprop functions to calculate the margin of error for a mean and a proportion. Functions are accessible from Data > Pivot and Data > Explore
+
+
+
+radiant.data 1.1.6
+- Improvements for wrapping generated code to Report > Rmd or Report > R
+
+-
+Data > Transform > Training now uses the
randomizr package to allow blocking variables when creating a training variables.
+
+
+radiant.data 1.1.3
+- Guard against using Data > Transform > Reorder/remove levels with too many levels (i.e., > 100)
+- Guard against using Data > Transform > Reorder/remove variables with too many variables (i.e., > 100)
+- Fix for DT table callbacks when shiny 1.4 hits CRAN (see https://github.com/rstudio/DT/issues/146#issuecomment-534319155)
+- Tables from Data > Pivot and Data > Explore now have
nr set to Inf by default (i.e., show all rows). The user can change this to the number of desired rows to show (e.g., select 3 rows in a sorted table)
+- Fix for example numbering in the help file for Data > Transform
+
+- Numerous small code changes to support enhanced auto-completion, tooltips, and annotations in shinyAce 0.4.1
+
+
+radiant.data 1.0.62019-08-22
+- Fix for
Data > Transform > Change type
+
+- Option to
fix_names to lower case
+- Keyboard shortcut (Enter) to load remove csv and rds files
+- Use a shinyAce input to generate data descriptions
+- Allow custom initial dataset list
+- Fix for latex formulas in Report > Rmd on Windows
+- Updated requirements for markdown and Rmarkdown
+- Fix for
radiant.init.data with shiny-server
+- Improvements to setup to allow access to server-side files by adding options to .Rprofile:
+
- Add
options(radiant.report = TRUE) to allow report generation in Report > Rmd and Report > R
+
+- Add
options(radiant.shinyFiles = TRUE) to allow server-side access to files
+- List specific directories you want to use with radiant using, for example,
options(radiant.sf_volumes = c(Git = "/home/jovyan/git"))
+
+
+
+
+radiant.data 1.0.02019-07-18
+- Support for series of class
ts (e.g., Data > Transform > Change type > Time series)
+- Require shinyAce 0.4.0
+- Vertical jitter set to 0 by default
+
+
+radiant.data 0.9.9.0
+- Added option to save Report > Rmd as a powerpoint file using
Rmarkdown
+
+- Removed dependency on
summarytools due to breaking changes
+- Fix for interaction (
iterm) and non-linear term (qterm) creation if character strings rather than integers are passed to the function
+- Remove specific symbols from reports in Report > Rmd to avoid issues when generating HTML or PDF documents
+- Keyboard shortcuts, i.e., CTRL-O and CTRL-S (CMD-O and CMD-S on macOS) to open and save data files in the Data > Manage tab
+- Various fixes to address breaking changes in dplyr 0.8.0
+- Added
radiant_ prefix to all attributes, except description, to avoid conflicts with other packages (e.g., vars in dplyr)
+
+
+radiant.data 0.9.8.6
+- Use
stringi::stri_trans_general to replace special symbols in Rmarkdown that may cause problems
+- Add empty line before and after code chunks when saving reports to Rmarkdown
+- Use
rio to load sav, dta, or sas7bdat files through the read files button in Report > Rmd and Report > R.
+- Create a
qscatter plot similar to the function of the same name in Stata
+- New radiant icon
+- Fix for setting where both
xlim and ylim are set in visualize function
+- Use an expandable
shinyAce input for the R-code log in Data > Transform
+
+
+
+radiant.data 0.9.8.0
+- Added an “autosave” options. Use
options(radiant.autosave = c(10, 180)); radiant::radiant() to auto-save the application state to the ~/.radiant.session folder every 10 minutes for the next 180 minutes. This can be useful if radiant is being used during an exam, for example.
+- Emergency backups are now saved to
~/.radiant.session/r_some_id.state.rda. The files should be automatically loaded when needed but can also be loaded as a regular radiant state file
+- Replace option to load an
.rda from from a URL in Data > Manage to load .rds files instead
+- Ensure variable and dataset names are valid for R (i.e., no spaces or symbols), “fixing” the input as needed
+- Fix to visualize now
ggplot::labs no longer accepts a list as input
+- Add option to generate square and cubed terms for use in linear and logistic regression in
radiant.model
+
+- Fix for error when trying to save invalid predictions in
radiant.model. This action now generates a pop-up in the browser interface
+- Add a specified description to a data.frame immediately on
register
+
+- Option to pass additional arguments to
shiny::runApp when starting radiant such as the port to use. For example, radiant.data::radiant.data(“https://github.com/radiant-rstats/docs/raw/gh-pages/examples/demo-dvd-rnd.state.rda”, port = 8080)
+- Option for automatic cleanup of deprecated code in both Report > Rmd and Report > R
+
+- Avoid attempt to fix deprecated code in Report > Rmd if
pred_data = ""
+
+- Fix for download icon linked to downloading of a state file after upgrade to shiny 1.2
+- Update documentation for Data > Combine
+
+- Fix for
format_df when the data.frame contains missing values. This fix is relevant for several summary functions run in Report > Rmd or Report > R
+
+- Fix for directory set when using
Knit report in Report > Rmd and Report > R without an Rstudio project. Will now correctly default to the working directory used in R(studio)
+- Added option to change
smooth setting for histograms with a density plot
+- Similar to
pmin and pmax, pfun et al. calculate summary statistics elementwise across multiple vectors
+- Add
Desktop as a default directory to show in the shinyFiles file browser
+- Load a state file on startup by providing a (relative) file path or a url. For example, radiant.data::radiant.data(“https://github.com/radiant-rstats/docs/raw/gh-pages/examples/demo-dvd-rnd.state.rda”) or radiant.data::radiant.data(“assignment.state.rda”)
+- Update example report in Report > Rmd
+
+- Add
deregister function to remove data in radiant from memory and the datasets dropdown list
+- Fix for invalid column names if used in
Data > Pivot
+
+
+
+radiant.data 0.9.7.0
+- Use
summarytools to generate summary information for datasets in Data > Manage
+
+- Show modal with warning about non-writable working directory when saving reports in Report > Rmd or Report > R
+
+- Apply
radiant.data::fix_names to files loaded into radiant to ensure valid R-object names
+- Use the content of the
Store filtered data as input to name the csv download in Data > View
+
+- Add “txt” as a recognized file type for
Read files in Report > Rmd and Report > R
+
+- Allow multiple
lines or loess curves based on a selected color variable for scatter plots in Data > Visualize
+
+- Indicate that a plot in Data > Visualize should be updated when plot labels are changed
+- Fix for #81 when variables used in Data > Pivot contain dots
+- Fix for
radiant.project_dir when no Rstudio project is used which could cause incorrect relative paths to be used
+- Fix code formatting for Report > Rmd when arguments include a list (e.g., ggplot labels)
+- On Linux use a modal to show code in Report > Rmd and Report > R when reporting is set to “manual”
+- Use
is_double to ensure dates are not treated as numeric variables in Data > View
+
+- Make sort and filter state of tables in Data > Explore and Data > Pivot available in Report > Rmd
+- Fix names for data sets loaded using the
Read files button in Report > Rmd or Report > R
+- Cleanup environment after closing app
+- Fix column names with spaces, etc. when reading csv files
+- Additional styling and labeling options for Data > Visualize are now available in the browser interface
+- Fix for code generation related to DT filters
+
+
+radiant.data 0.9.6.14
+
+Major changes
+- Using
shinyFiles to provide convenient access to data located on a server
+- Avoid
XQuartz requirement
+
+
+Minor changes
+- Load
data(...) into the current environment rather than defaulting only to the global environment
+-
+
file.rename failed using docker on windows when saving a report. Using file.copy instead
+- Fix for
sf_volumes used to set the root directories to load and save files
+- Set default locale to “en_US.UTF-8” when using shiny-server unless
Sys.getlocale(category = "LC_ALL") what set to something other than “C”
+- Modal shown if and Rmd (R) file is not available when using “To Rstudio (Rmd)” in Report > Rmd or “To Rstudio (R)” in Report > R
+
+- Track progress loading (state) files
+- Fix for
radiant.sf_volumes used for the shinyFiles file browser
+- Improvements for sending code from Radiant to Rstudio
+- Better support for paths when using radiant on a server (i.e., revert to home directory using
radiant.data::find_home())
+- Revert from
svg to png for plots in _Report > Rmd_ and _Report > R_.svg` scatter plots with many point get to big for practical use on servers that have to transfer images to a local browser
+- Removed dependency on
methods package
+
+
+
+radiant.data 0.9.5.3
+- Fix smart comma’s in data descriptions
+- Search and replace
desc(n) in reports and replace by desc(n_obs)
+
+- Revert to storing the r_data environment as a list on stop to avoid reference problems (@josh1400)
+- Fix for plot type in Data > Pivot in older state files (@josh1400)
+- Used all declared imports (CRAN)
+
+
+radiant.data 0.9.5.0
+- Fix for
radiant.data::explore when variable names contain an underscore
+- Fix for
find_gdrive when drive is not being synced
+- Fixes in Report > Rmd and Report > R to accommodate for pandoc > 2
+
+
+radiant.data 0.9.4.6
+- Don’t update a reactive binding for an object if the binding already exists. See issue https://github.com/rstudio/shiny/issues/2065
+
+- Fix to accommodate changes in
deparse in R 3.5
+- Fix for saving data in Data > Manage and generating the relevant R-code
+
+
+
+
+radiant.data 0.9.3.3
+
+Major changes
+- When using radiant with Rstudio Viewer or in an Rstudio Window, loading and saving data through Data > Manage generates R-code the user can add to Report > Rmd or Report > R. Clicking the
Show R-code checkbox displays the R-code used to load or save the current dataset
+- Various changes to the code to accommodate the use of
shiny::makeReactiveBinding. The advantage is that the code generated for Report > Rmd and Report > R will no longer have to use a list (r_data) to store and access data. This means that code generated and used in the Radiant browser interface will be directly usable without the browser interface as well
+- Removed
loadr, saver, load_csv, loadcsv_url, loadrds_url, and make_funs functions as they are no longer needed
+- Deprecated
mean_rm, median_rm, min_rm, max_rm,sd_rm,var_rm, and sum_rm functions as they are no longer needed
+
+
+Minor changes
+- Added
load_clip and save_clip to load and save data to the clipboard on Windows and macOS
+- Improved auto completion in Report > Rmd and Report > R
+
+- Maintain, store, and clean the settings of the interactive table in Data > View
+
+- Address closing Rstudio Window issue (https://github.com/rstudio/shiny/issues/2033)
+
+
+
+radiant.data 0.9.2.3
+
+Major changes
+-
+Report > Rmd and Report > R will now be evaluated the
r_data environment. This means that the return value from ls() will be much cleaner
+
+
+Minor changes
+- Add option to load files with extension .rdata or .tsv using
loadr which add that data to the Datasets dropdown
+-
+
visualize will default to a scatter plot if xvar and yvar are specified but no plot type is provided in the function call
+- Improvements to
read_files function to interactively generate R-code (or Rmarkdown code-chunks) to read files in various format (e.g., SQLite, rds, csv, xlsx, css, jpg, etc.). Supports relative paths and uses find_dropbox() and find_gdrive() when applicable
+
+
+
+radiant.data 0.9.2.2
+
+Minor changes
+- Require
shinyAce 0.3.0
+- Export
read_files function to interactively generate R-code or Rmarkdown code-chunks to read files in various format (e.g., SQLite, rds, csv, xlsx, css, jpg, etc.). Supports relative paths and uses find_dropbox() and find_gdrive() when applicable
+
+
+
+
+CHANGES IN radiant.data 0.9.0.22
+
+Bug fixes
+- Fix for #43 where scatter plot was not shown for a dataset with less than 1,000 rows
+- Fix for Report > Rmd and Report > R when R-code or Rmarkdown is being pulled from the Rstudio editor
+
+
+
+
+
+radiant.data 0.9.0.16
+
+Minor changes
+- Changed license for help files and images for radiant.data to CC-BY-SA
+
+
+
+
+radiant.data 0.9.0.15
+
+
+
+
+radiant.data 0.9.0.13
+
+Minor changes
+- Apply
fixMS to replace curly quotes, em dash, etc. when using Data > Transform > Create
+
+- Option to set number of decimals to show in Data > View
+
+- Improved number formatting in interactive tables in Data > View, Data > Pivot, and Data > Explore
+
+- Option to include an interactive view of a dataset in Report > Rmd. By default, the number of rows is set to 100 as, most likely, the user will not want to embed a large dataset in save HTML report
+-
+Data > Transform will leave variables selected, unless switching to
Create or Spread
+
+- Switch focus to editor in Report > Rmd and Report > R when no other input has focus
+
+
+
+
+radiant.data 0.9.0.7
+
+Minor changes
+- Allow response variables with NA values in Model > Logistic regression and other classification models
+- Support logicals in code generation from Data > View
+
+- Track window size using
input$get_screen_width
+
+- Focus on editor when switching to Report > Rmd or Report > R so generated code is shown immediately and the user can navigate and type in the editor without having to click first
+- Add information about the first level when plotting a bar chart with a categorical variable on the Y-axis (e.g., mean(buyer {yes}))
+
+
+Bug fixes
+- Cleanup now also occurs when the stop button is used in Rstudio to close the app
+- Fix to include
DiagrammeR based plots in Rmarkdown reports
+- Fix in
read_files for SQLite data names
+- De-activate spell check auto correction in
selectizeInput in Rstudio Viewer shiny #1916
+
+- Fix to allow selecting and copying text output from Report > Rmd and Report > R
+
+- Remove “fancy” quotes from filters
+- Known issue: The Rstudio viewer may not always close the viewer window when trying to stop the application with the
Stop link in the navbar. As a work-around, use Rstudio’s stop buttons instead.
+
+
+
+radiant.data 0.9.0.0
+
+Major changes
+- If Rstudio project is used Report > Rmd and Report > R will use the project directory as base. This allows users to use relative paths and making it easier to share (reproducible) code
+- Specify options in .Rprofile for upload memory limit and running Report > Rmd on server
+-
+
find_project function based on rstudioapi
+
+-
+Report > Rmd Read button to generate code to load various types of data (e.g., rda, rds, xls, yaml, feather)
+-
+Report > Rmd Read button to generate code to load various types of files in report (e.g., jpg, png, md, Rmd, R). If Radiant was started from an Rstudio project, the file paths used will be relative to the project root. Paths to files synced to local Dropbox or Google Drive folder will use the
find_dropbox and find_gdrive functions to enhances reproducibility.
+-
+Report > Rmd Load Report button can be used to load Rmarkdown file in the editor. It will also extract the source code from Notebook and HTML files with embedded Rmarkdown
+-
+Report > Rmd will read Rmd directly from Rstudio when “To Rstudio (Rmd)” is selected. This will make it possible to use Rstudio Server Pro’s Share project option for realtime collaboration in Radiant
+- Long lines of code generated for Report > Rmd will be wrapped to enhance readability
+-
+Report > R is now equivalent to Report > Rmd but in R-code format
+-
+Report > Rmd option to view Editor, Preview, or Both
+- Show Rstudio project information in navbar if available
+
+
+
+
+radiant.data 0.8.7.8
+
+Minor changes
+- Added preview options to Data > Manage based on https://github.com/radiant-rstats/radiant/issues/30
+
+- Add selected dataset name as default table download name in Data > View, Data > Pivot, and Data > Explore
+
+- Use “stack” as the default for histograms and frequency charts in Data > Visualize
+
+- Cleanup
Stop & Report option in navbar
+- Upgraded tidyr dependency to 0.7
+- Upgraded dplyr dependency to 0.7.1
+
+
+
+
+radiant.data 0.8.6.0
+
+Minor changes
+- Export
ggplotly from plotly for interactive plots in Report > Rmd
+
+- Export
subplot from plotly for grids of interactive plots in Report > Rmd
+
+- Set default
res = 96 for renderPlot and dpi = 96 for knitr::opts_chunk
+
+- Add
fillcol, linecol, and pointcol to visualize to set plot colors when no fill or color variable has been selected
+- Reverse legend ordering in Data > Visualize when axes are flipped using
coor_flip()
+
+- Added functions to choose.files and choose.dir. Uses JavaScript on Mac, utils::choose.files and utils::choose.dir on Windows, and reverts to file.choose on Linux
+- Added
find_gdrive to determine the path to a user’s local Google Drive folder if available
+-
+
fixMs for encoding in reports on Windows
+
+
+
+
+radiant.data 0.8.1.0
+
+Minor changes
+- Specify the maximum number of rows to load for a csv and csv (url) file through Data > Manage
+
+- Support for loading and saving feather files, including specifying the maximum number of rows to load through Data > Manage
+
+- Added author and year arguments to help modals in inst/app/radiant.R (thanks @kmezhoud)
+- Added size argument for scatter plots to create bubble charts (thanks @andrewsali)
+- Example and CSS formatting for tables in Report > Rmd
+
+- Added
seed argument to make_train
+
+- Added
prop, sdprop, etc. for working with proportions
+- Set
ylim in visualize for multiple plots
+- Show progress indicator when saving reports from Report > Rmd
+
+-
+
copy_attr convenience function
+-
+
refactor function to keep only a subset of levels in a factor and recode the remaining (and first) level to, for example, other
+-
+
register function to add a (transformed) dataset to the dataset dropdown
+- Remember name of state files loaded and suggest that name when re-saving the state
+- Show dataset name in output if dataframe passed directly to analysis function
+- R-notebooks are now the default option for output saved from Report > Rmd and Report > R
+
+- Improved documentation on how to customize plots in Report > Rmd
+
+- Keyboard short-cut to put code into Report > Rmd (ALT-enter)
+
+
+Bug fixes
+- When clicking the
rename button, without changing the name, the dataset was set to NULL (thanks @kmezhoud, https://github.com/radiant-rstats/radiant/issues/5)
+- Replace ext with .ext in
mutate_each function call
+- Variance estimation in Data > Explore would cause an error with unit cell-frequencies (thanks @kmezhoud, https://github.com/radiant-rstats/radiant/issues/6)
+- Fix for as_integer when factor levels are characters
+- Fix for integer conversion in explore
+- Remove \r and special characters from strings in r_data and r_state
+- Fix sorting in Report > Rmd for tables created using Data > Pivot and Data > Explore when column headers contain symbols or spaces (thanks @4kammer)
+- Set
error = TRUE for rmarkdown for consistency with knitr as used in Report > Rmd
+
+- Correctly handle decimal indicators when loading csv files in Data > Manage
+
+- Don’t overwrite a dataset to combine if combine generates an error when user sets the the name of the combined data to that of an already selected dataset
+- When multiple variables were selected, data were not correctly summarized in Data > Transform
+- Add (function) label to bar plot when x-variable is an integer
+- Maintain order of variables in Data > Visualize when using “color”, “fill”, “comby”, or “combx”
+- Avoid warning when switching datasets in Data > Transform and variables being summarized do not exists in the new dataset
+- which.pmax produced a list but needed to be integer
+- To customized predictions in radiant.model indexr must be able to customize the prediction dataframe
+- describe now correctly resets the working directory on exit
+- removed all calls to summarise_each and mutate_each from dplyr
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/radiant.data/docs/pkgdown.css b/radiant.data/docs/pkgdown.css
new file mode 100644
index 0000000000000000000000000000000000000000..80ea5b838a4d8f24ae023bbf3582a8079aadbf66
--- /dev/null
+++ b/radiant.data/docs/pkgdown.css
@@ -0,0 +1,384 @@
+/* Sticky footer */
+
+/**
+ * Basic idea: https://philipwalton.github.io/solved-by-flexbox/demos/sticky-footer/
+ * Details: https://github.com/philipwalton/solved-by-flexbox/blob/master/assets/css/components/site.css
+ *
+ * .Site -> body > .container
+ * .Site-content -> body > .container .row
+ * .footer -> footer
+ *
+ * Key idea seems to be to ensure that .container and __all its parents__
+ * have height set to 100%
+ *
+ */
+
+html, body {
+ height: 100%;
+}
+
+body {
+ position: relative;
+}
+
+body > .container {
+ display: flex;
+ height: 100%;
+ flex-direction: column;
+}
+
+body > .container .row {
+ flex: 1 0 auto;
+}
+
+footer {
+ margin-top: 45px;
+ padding: 35px 0 36px;
+ border-top: 1px solid #e5e5e5;
+ color: #666;
+ display: flex;
+ flex-shrink: 0;
+}
+footer p {
+ margin-bottom: 0;
+}
+footer div {
+ flex: 1;
+}
+footer .pkgdown {
+ text-align: right;
+}
+footer p {
+ margin-bottom: 0;
+}
+
+img.icon {
+ float: right;
+}
+
+/* Ensure in-page images don't run outside their container */
+.contents img {
+ max-width: 100%;
+ height: auto;
+}
+
+/* Fix bug in bootstrap (only seen in firefox) */
+summary {
+ display: list-item;
+}
+
+/* Typographic tweaking ---------------------------------*/
+
+.contents .page-header {
+ margin-top: calc(-60px + 1em);
+}
+
+dd {
+ margin-left: 3em;
+}
+
+/* Section anchors ---------------------------------*/
+
+a.anchor {
+ display: none;
+ margin-left: 5px;
+ width: 20px;
+ height: 20px;
+
+ background-image: url(./link.svg);
+ background-repeat: no-repeat;
+ background-size: 20px 20px;
+ background-position: center center;
+}
+
+h1:hover .anchor,
+h2:hover .anchor,
+h3:hover .anchor,
+h4:hover .anchor,
+h5:hover .anchor,
+h6:hover .anchor {
+ display: inline-block;
+}
+
+/* Fixes for fixed navbar --------------------------*/
+
+.contents h1, .contents h2, .contents h3, .contents h4 {
+ padding-top: 60px;
+ margin-top: -40px;
+}
+
+/* Navbar submenu --------------------------*/
+
+.dropdown-submenu {
+ position: relative;
+}
+
+.dropdown-submenu>.dropdown-menu {
+ top: 0;
+ left: 100%;
+ margin-top: -6px;
+ margin-left: -1px;
+ border-radius: 0 6px 6px 6px;
+}
+
+.dropdown-submenu:hover>.dropdown-menu {
+ display: block;
+}
+
+.dropdown-submenu>a:after {
+ display: block;
+ content: " ";
+ float: right;
+ width: 0;
+ height: 0;
+ border-color: transparent;
+ border-style: solid;
+ border-width: 5px 0 5px 5px;
+ border-left-color: #cccccc;
+ margin-top: 5px;
+ margin-right: -10px;
+}
+
+.dropdown-submenu:hover>a:after {
+ border-left-color: #ffffff;
+}
+
+.dropdown-submenu.pull-left {
+ float: none;
+}
+
+.dropdown-submenu.pull-left>.dropdown-menu {
+ left: -100%;
+ margin-left: 10px;
+ border-radius: 6px 0 6px 6px;
+}
+
+/* Sidebar --------------------------*/
+
+#pkgdown-sidebar {
+ margin-top: 30px;
+ position: -webkit-sticky;
+ position: sticky;
+ top: 70px;
+}
+
+#pkgdown-sidebar h2 {
+ font-size: 1.5em;
+ margin-top: 1em;
+}
+
+#pkgdown-sidebar h2:first-child {
+ margin-top: 0;
+}
+
+#pkgdown-sidebar .list-unstyled li {
+ margin-bottom: 0.5em;
+}
+
+/* bootstrap-toc tweaks ------------------------------------------------------*/
+
+/* All levels of nav */
+
+nav[data-toggle='toc'] .nav > li > a {
+ padding: 4px 20px 4px 6px;
+ font-size: 1.5rem;
+ font-weight: 400;
+ color: inherit;
+}
+
+nav[data-toggle='toc'] .nav > li > a:hover,
+nav[data-toggle='toc'] .nav > li > a:focus {
+ padding-left: 5px;
+ color: inherit;
+ border-left: 1px solid #878787;
+}
+
+nav[data-toggle='toc'] .nav > .active > a,
+nav[data-toggle='toc'] .nav > .active:hover > a,
+nav[data-toggle='toc'] .nav > .active:focus > a {
+ padding-left: 5px;
+ font-size: 1.5rem;
+ font-weight: 400;
+ color: inherit;
+ border-left: 2px solid #878787;
+}
+
+/* Nav: second level (shown on .active) */
+
+nav[data-toggle='toc'] .nav .nav {
+ display: none; /* Hide by default, but at >768px, show it */
+ padding-bottom: 10px;
+}
+
+nav[data-toggle='toc'] .nav .nav > li > a {
+ padding-left: 16px;
+ font-size: 1.35rem;
+}
+
+nav[data-toggle='toc'] .nav .nav > li > a:hover,
+nav[data-toggle='toc'] .nav .nav > li > a:focus {
+ padding-left: 15px;
+}
+
+nav[data-toggle='toc'] .nav .nav > .active > a,
+nav[data-toggle='toc'] .nav .nav > .active:hover > a,
+nav[data-toggle='toc'] .nav .nav > .active:focus > a {
+ padding-left: 15px;
+ font-weight: 500;
+ font-size: 1.35rem;
+}
+
+/* orcid ------------------------------------------------------------------- */
+
+.orcid {
+ font-size: 16px;
+ color: #A6CE39;
+ /* margins are required by official ORCID trademark and display guidelines */
+ margin-left:4px;
+ margin-right:4px;
+ vertical-align: middle;
+}
+
+/* Reference index & topics ----------------------------------------------- */
+
+.ref-index th {font-weight: normal;}
+
+.ref-index td {vertical-align: top; min-width: 100px}
+.ref-index .icon {width: 40px;}
+.ref-index .alias {width: 40%;}
+.ref-index-icons .alias {width: calc(40% - 40px);}
+.ref-index .title {width: 60%;}
+
+.ref-arguments th {text-align: right; padding-right: 10px;}
+.ref-arguments th, .ref-arguments td {vertical-align: top; min-width: 100px}
+.ref-arguments .name {width: 20%;}
+.ref-arguments .desc {width: 80%;}
+
+/* Nice scrolling for wide elements --------------------------------------- */
+
+table {
+ display: block;
+ overflow: auto;
+}
+
+/* Syntax highlighting ---------------------------------------------------- */
+
+pre, code, pre code {
+ background-color: #f8f8f8;
+ color: #333;
+}
+pre, pre code {
+ white-space: pre-wrap;
+ word-break: break-all;
+ overflow-wrap: break-word;
+}
+
+pre {
+ border: 1px solid #eee;
+}
+
+pre .img, pre .r-plt {
+ margin: 5px 0;
+}
+
+pre .img img, pre .r-plt img {
+ background-color: #fff;
+}
+
+code a, pre a {
+ color: #375f84;
+}
+
+a.sourceLine:hover {
+ text-decoration: none;
+}
+
+.fl {color: #1514b5;}
+.fu {color: #000000;} /* function */
+.ch,.st {color: #036a07;} /* string */
+.kw {color: #264D66;} /* keyword */
+.co {color: #888888;} /* comment */
+
+.error {font-weight: bolder;}
+.warning {font-weight: bolder;}
+
+/* Clipboard --------------------------*/
+
+.hasCopyButton {
+ position: relative;
+}
+
+.btn-copy-ex {
+ position: absolute;
+ right: 0;
+ top: 0;
+ visibility: hidden;
+}
+
+.hasCopyButton:hover button.btn-copy-ex {
+ visibility: visible;
+}
+
+/* headroom.js ------------------------ */
+
+.headroom {
+ will-change: transform;
+ transition: transform 200ms linear;
+}
+.headroom--pinned {
+ transform: translateY(0%);
+}
+.headroom--unpinned {
+ transform: translateY(-100%);
+}
+
+/* mark.js ----------------------------*/
+
+mark {
+ background-color: rgba(255, 255, 51, 0.5);
+ border-bottom: 2px solid rgba(255, 153, 51, 0.3);
+ padding: 1px;
+}
+
+/* vertical spacing after htmlwidgets */
+.html-widget {
+ margin-bottom: 10px;
+}
+
+/* fontawesome ------------------------ */
+
+.fab {
+ font-family: "Font Awesome 5 Brands" !important;
+}
+
+/* don't display links in code chunks when printing */
+/* source: https://stackoverflow.com/a/10781533 */
+@media print {
+ code a:link:after, code a:visited:after {
+ content: "";
+ }
+}
+
+/* Section anchors ---------------------------------
+ Added in pandoc 2.11: https://github.com/jgm/pandoc-templates/commit/9904bf71
+*/
+
+div.csl-bib-body { }
+div.csl-entry {
+ clear: both;
+}
+.hanging-indent div.csl-entry {
+ margin-left:2em;
+ text-indent:-2em;
+}
+div.csl-left-margin {
+ min-width:2em;
+ float:left;
+}
+div.csl-right-inline {
+ margin-left:2em;
+ padding-left:1em;
+}
+div.csl-indent {
+ margin-left: 2em;
+}
diff --git a/radiant.data/docs/pkgdown.js b/radiant.data/docs/pkgdown.js
new file mode 100644
index 0000000000000000000000000000000000000000..6f0eee40bc3c69061d33fbed1bc9644f47c0eb8a
--- /dev/null
+++ b/radiant.data/docs/pkgdown.js
@@ -0,0 +1,108 @@
+/* http://gregfranko.com/blog/jquery-best-practices/ */
+(function($) {
+ $(function() {
+
+ $('.navbar-fixed-top').headroom();
+
+ $('body').css('padding-top', $('.navbar').height() + 10);
+ $(window).resize(function(){
+ $('body').css('padding-top', $('.navbar').height() + 10);
+ });
+
+ $('[data-toggle="tooltip"]').tooltip();
+
+ var cur_path = paths(location.pathname);
+ var links = $("#navbar ul li a");
+ var max_length = -1;
+ var pos = -1;
+ for (var i = 0; i < links.length; i++) {
+ if (links[i].getAttribute("href") === "#")
+ continue;
+ // Ignore external links
+ if (links[i].host !== location.host)
+ continue;
+
+ var nav_path = paths(links[i].pathname);
+
+ var length = prefix_length(nav_path, cur_path);
+ if (length > max_length) {
+ max_length = length;
+ pos = i;
+ }
+ }
+
+ // Add class to parent
`).
+ getTopLevel: function($scope) {
+ for (var i = 1; i <= 6; i++) {
+ var $headings = this.findOrFilter($scope, 'h' + i);
+ if ($headings.length > 1) {
+ return i;
+ }
+ }
+
+ return 1;
+ },
+
+ // returns the elements for the top level, and the next below it
+ getHeadings: function($scope, topLevel) {
+ var topSelector = 'h' + topLevel;
+
+ var secondaryLevel = topLevel + 1;
+ var secondarySelector = 'h' + secondaryLevel;
+
+ return this.findOrFilter($scope, topSelector + ',' + secondarySelector);
+ },
+
+ getNavLevel: function(el) {
+ return parseInt(el.tagName.charAt(1), 10);
+ },
+
+ populateNav: function($topContext, topLevel, $headings) {
+ var $context = $topContext;
+ var $prevNav;
+
+ var helpers = this;
+ $headings.each(function(i, el) {
+ var $newNav = helpers.generateNavItem(el);
+ var navLevel = helpers.getNavLevel(el);
+
+ // determine the proper $context
+ if (navLevel === topLevel) {
+ // use top level
+ $context = $topContext;
+ } else if ($prevNav && $context === $topContext) {
+ // create a new level of the tree and switch to it
+ $context = helpers.createChildNavList($prevNav);
+ } // else use the current $context
+
+ $context.append($newNav);
+
+ $prevNav = $newNav;
+ });
+ },
+
+ parseOps: function(arg) {
+ var opts;
+ if (arg.jquery) {
+ opts = {
+ $nav: arg
+ };
+ } else {
+ opts = arg;
+ }
+ opts.$scope = opts.$scope || $(document.body);
+ return opts;
+ }
+ },
+
+ // accepts a jQuery object, or an options object
+ init: function(opts) {
+ opts = this.helpers.parseOps(opts);
+
+ // ensure that the data attribute is in place for styling
+ opts.$nav.attr('data-toggle', 'toc');
+
+ var $topContext = this.helpers.createChildNavList(opts.$nav);
+ var topLevel = this.helpers.getTopLevel(opts.$scope);
+ var $headings = this.helpers.getHeadings(opts.$scope, topLevel);
+ this.helpers.populateNav($topContext, topLevel, $headings);
+ }
+ };
+
+ $(function() {
+ $('nav[data-toggle="toc"]').each(function(i, el) {
+ var $nav = $(el);
+ Toc.init($nav);
+ });
+ });
+})();
diff --git a/radiant.data/docs/docsearch.css b/radiant.data/docs/docsearch.css
new file mode 100644
index 0000000000000000000000000000000000000000..e5f1fe1dfa2c34c51fe941829b511acd8c763301
--- /dev/null
+++ b/radiant.data/docs/docsearch.css
@@ -0,0 +1,148 @@
+/* Docsearch -------------------------------------------------------------- */
+/*
+ Source: https://github.com/algolia/docsearch/
+ License: MIT
+*/
+
+.algolia-autocomplete {
+ display: block;
+ -webkit-box-flex: 1;
+ -ms-flex: 1;
+ flex: 1
+}
+
+.algolia-autocomplete .ds-dropdown-menu {
+ width: 100%;
+ min-width: none;
+ max-width: none;
+ padding: .75rem 0;
+ background-color: #fff;
+ background-clip: padding-box;
+ border: 1px solid rgba(0, 0, 0, .1);
+ box-shadow: 0 .5rem 1rem rgba(0, 0, 0, .175);
+}
+
+@media (min-width:768px) {
+ .algolia-autocomplete .ds-dropdown-menu {
+ width: 175%
+ }
+}
+
+.algolia-autocomplete .ds-dropdown-menu::before {
+ display: none
+}
+
+.algolia-autocomplete .ds-dropdown-menu [class^=ds-dataset-] {
+ padding: 0;
+ background-color: rgb(255,255,255);
+ border: 0;
+ max-height: 80vh;
+}
+
+.algolia-autocomplete .ds-dropdown-menu .ds-suggestions {
+ margin-top: 0
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion {
+ padding: 0;
+ overflow: visible
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--category-header {
+ padding: .125rem 1rem;
+ margin-top: 0;
+ font-size: 1.3em;
+ font-weight: 500;
+ color: #00008B;
+ border-bottom: 0
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--wrapper {
+ float: none;
+ padding-top: 0
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--subcategory-column {
+ float: none;
+ width: auto;
+ padding: 0;
+ text-align: left
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--content {
+ float: none;
+ width: auto;
+ padding: 0
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--content::before {
+ display: none
+}
+
+.algolia-autocomplete .ds-suggestion:not(:first-child) .algolia-docsearch-suggestion--category-header {
+ padding-top: .75rem;
+ margin-top: .75rem;
+ border-top: 1px solid rgba(0, 0, 0, .1)
+}
+
+.algolia-autocomplete .ds-suggestion .algolia-docsearch-suggestion--subcategory-column {
+ display: block;
+ padding: .1rem 1rem;
+ margin-bottom: 0.1;
+ font-size: 1.0em;
+ font-weight: 400
+ /* display: none */
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--title {
+ display: block;
+ padding: .25rem 1rem;
+ margin-bottom: 0;
+ font-size: 0.9em;
+ font-weight: 400
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--text {
+ padding: 0 1rem .5rem;
+ margin-top: -.25rem;
+ font-size: 0.8em;
+ font-weight: 400;
+ line-height: 1.25
+}
+
+.algolia-autocomplete .algolia-docsearch-footer {
+ width: 110px;
+ height: 20px;
+ z-index: 3;
+ margin-top: 10.66667px;
+ float: right;
+ font-size: 0;
+ line-height: 0;
+}
+
+.algolia-autocomplete .algolia-docsearch-footer--logo {
+ background-image: url("data:image/svg+xml;utf8,");
+ background-repeat: no-repeat;
+ background-position: 50%;
+ background-size: 100%;
+ overflow: hidden;
+ text-indent: -9000px;
+ width: 100%;
+ height: 100%;
+ display: block;
+ transform: translate(-8px);
+}
+
+.algolia-autocomplete .algolia-docsearch-suggestion--highlight {
+ color: #FF8C00;
+ background: rgba(232, 189, 54, 0.1)
+}
+
+
+.algolia-autocomplete .algolia-docsearch-suggestion--text .algolia-docsearch-suggestion--highlight {
+ box-shadow: inset 0 -2px 0 0 rgba(105, 105, 105, .5)
+}
+
+.algolia-autocomplete .ds-suggestion.ds-cursor .algolia-docsearch-suggestion--content {
+ background-color: rgba(192, 192, 192, .15)
+}
diff --git a/radiant.data/docs/docsearch.js b/radiant.data/docs/docsearch.js
new file mode 100644
index 0000000000000000000000000000000000000000..b35504cd3a282816130a16881f3ebeead9c1bcb4
--- /dev/null
+++ b/radiant.data/docs/docsearch.js
@@ -0,0 +1,85 @@
+$(function() {
+
+ // register a handler to move the focus to the search bar
+ // upon pressing shift + "/" (i.e. "?")
+ $(document).on('keydown', function(e) {
+ if (e.shiftKey && e.keyCode == 191) {
+ e.preventDefault();
+ $("#search-input").focus();
+ }
+ });
+
+ $(document).ready(function() {
+ // do keyword highlighting
+ /* modified from https://jsfiddle.net/julmot/bL6bb5oo/ */
+ var mark = function() {
+
+ var referrer = document.URL ;
+ var paramKey = "q" ;
+
+ if (referrer.indexOf("?") !== -1) {
+ var qs = referrer.substr(referrer.indexOf('?') + 1);
+ var qs_noanchor = qs.split('#')[0];
+ var qsa = qs_noanchor.split('&');
+ var keyword = "";
+
+ for (var i = 0; i < qsa.length; i++) {
+ var currentParam = qsa[i].split('=');
+
+ if (currentParam.length !== 2) {
+ continue;
+ }
+
+ if (currentParam[0] == paramKey) {
+ keyword = decodeURIComponent(currentParam[1].replace(/\+/g, "%20"));
+ }
+ }
+
+ if (keyword !== "") {
+ $(".contents").unmark({
+ done: function() {
+ $(".contents").mark(keyword);
+ }
+ });
+ }
+ }
+ };
+
+ mark();
+ });
+});
+
+/* Search term highlighting ------------------------------*/
+
+function matchedWords(hit) {
+ var words = [];
+
+ var hierarchy = hit._highlightResult.hierarchy;
+ // loop to fetch from lvl0, lvl1, etc.
+ for (var idx in hierarchy) {
+ words = words.concat(hierarchy[idx].matchedWords);
+ }
+
+ var content = hit._highlightResult.content;
+ if (content) {
+ words = words.concat(content.matchedWords);
+ }
+
+ // return unique words
+ var words_uniq = [...new Set(words)];
+ return words_uniq;
+}
+
+function updateHitURL(hit) {
+
+ var words = matchedWords(hit);
+ var url = "";
+
+ if (hit.anchor) {
+ url = hit.url_without_anchor + '?q=' + escape(words.join(" ")) + '#' + hit.anchor;
+ } else {
+ url = hit.url + '?q=' + escape(words.join(" "));
+ }
+
+ return url;
+}
diff --git a/radiant.data/docs/docsearch.json b/radiant.data/docs/docsearch.json
new file mode 100644
index 0000000000000000000000000000000000000000..715937648ea49c8f2e3f10d27fa5c14b75d844a8
--- /dev/null
+++ b/radiant.data/docs/docsearch.json
@@ -0,0 +1,95 @@
+{
+ "index_name": "radiant_data",
+ "start_urls": [
+ {
+ "url": "https://radiant-rstats.github.io/radiant.data/index.html",
+ "selectors_key": "homepage",
+ "tags": [
+ "homepage"
+ ]
+ },
+ {
+ "url": "https://radiant-rstats.github.io/radiant.data/reference",
+ "selectors_key": "reference",
+ "tags": [
+ "reference"
+ ]
+ },
+ {
+ "url": "https://radiant-rstats.github.io/radiant.data/articles",
+ "selectors_key": "articles",
+ "tags": [
+ "articles"
+ ]
+ }
+ ],
+ "stop_urls": [
+ "/reference/$",
+ "/reference/index.html",
+ "/articles/$",
+ "/articles/index.html"
+ ],
+ "sitemap_urls": [
+ "https://radiant-rstats.github.io/radiant.data/sitemap.xml"
+ ],
+ "selectors": {
+ "homepage": {
+ "lvl0": {
+ "selector": ".contents h1",
+ "default_value": "radiant.data Home page"
+ },
+ "lvl1": {
+ "selector": ".contents h2"
+ },
+ "lvl2": {
+ "selector": ".contents h3",
+ "default_value": "Context"
+ },
+ "lvl3": ".ref-arguments td, .ref-description",
+ "text": ".contents p, .contents li, .contents .pre"
+ },
+ "reference": {
+ "lvl0": {
+ "selector": ".contents h1"
+ },
+ "lvl1": {
+ "selector": ".contents .name",
+ "default_value": "Argument"
+ },
+ "lvl2": {
+ "selector": ".ref-arguments th",
+ "default_value": "Description"
+ },
+ "lvl3": ".ref-arguments td, .ref-description",
+ "text": ".contents p, .contents li"
+ },
+ "articles": {
+ "lvl0": {
+ "selector": ".contents h1"
+ },
+ "lvl1": {
+ "selector": ".contents .name"
+ },
+ "lvl2": {
+ "selector": ".contents h2, .contents h3",
+ "default_value": "Context"
+ },
+ "text": ".contents p, .contents li"
+ }
+ },
+ "selectors_exclude": [
+ ".dont-index"
+ ],
+ "min_indexed_level": 2,
+ "custom_settings": {
+ "separatorsToIndex": "_",
+ "attributesToRetrieve": [
+ "hierarchy",
+ "content",
+ "anchor",
+ "url",
+ "url_without_anchor"
+ ]
+ }
+}
+
diff --git a/radiant.data/docs/index.html b/radiant.data/docs/index.html
new file mode 100644
index 0000000000000000000000000000000000000000..3c25a5ddaedeb3ee2c75f8e391faa7ade42fa79d
--- /dev/null
+++ b/radiant.data/docs/index.html
@@ -0,0 +1,365 @@
+
+
+
+
+
+
+
+Data Menu for Radiant: Business Analytics using R and Shiny • radiant.data
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+Radiant is an open-source platform-independent browser-based interface for business analytics in R. The application is based on the Shiny package and can be run locally or on a server. Radiant was developed by Vincent Nijs. Please use the issue tracker on GitHub to suggest enhancements or report problems: https://github.com/radiant-rstats/radiant.data/issues. For other questions and comments please use radiant@rady.ucsd.edu.
+
+Key features
+
+
+- Explore: Quickly and easily summarize, visualize, and analyze your data
+- Cross-platform: It runs in a browser on Windows, Mac, and Linux
+- Reproducible: Recreate results and share work with others as a state file or an Rmarkdown report
+- Programming: Integrate Radiant’s analysis functions with your own R-code
+- Context: Data and examples focus on business applications
+
+
+
+Playlists
+
+There are two youtube playlists with video tutorials. The first provides a general introduction to key features in Radiant. The second covers topics relevant in a course on business analytics (i.e., Probability, Decision Analysis, Hypothesis Testing, Linear Regression, and Simulation).
+
+
+
+Explore
+
+Radiant is interactive. Results update immediately when inputs are changed (i.e., no separate dialog boxes) and/or when a button is pressed (e.g., Estimate in Model > Estimate > Logistic regression (GLM)). This facilitates rapid exploration and understanding of the data.
+
+
+Cross-platform
+
+Radiant works on Windows, Mac, or Linux. It can run without an Internet connection and no data will leave your computer. You can also run the app as a web application on a server.
+
+
+Reproducible
+
+To conduct high-quality analysis, simply saving output is not enough. You need the ability to reproduce results for the same data and/or when new data become available. Moreover, others may want to review your analysis and results. Save and load the state of the application to continue your work at a later time or on another computer. Share state files with others and create reproducible reports using Rmarkdown. See also the section on Saving and loading state below
+If you are using Radiant on a server you can even share the URL (include the SSUID) with others so they can see what you are working on. Thanks for this feature go to Joe Cheng.
+
+
+Programming
+
+Although Radiant’s web-interface can handle quite a few data and analysis tasks, you may prefer to write your own R-code. Radiant provides a bridge to programming in R(studio) by exporting the functions used for analysis (i.e., you can conduct your analysis using the Radiant web-interface or by calling Radiant’s functions directly from R-code). For more information about programming with Radiant see the programming page on the documentation site.
+
+
+
+
+How to install Radiant
+
+
+In Rstudio you can start and update Radiant through the Addins menu at the top of the screen. To install the latest version of Radiant for Windows or Mac, with complete documentation for off-line access, open R(studio) and copy-and-paste the command below:
+
+options(repos = c(RSM = "https://radiant-rstats.github.io/minicran", CRAN = "https://cloud.r-project.org"))
+install.packages("radiant")
+Once all packages are installed, select Start radiant from the Addins menu in Rstudio or use the command below to launch the app:
+
+radiant::radiant()
+To launch Radiant in Rstudio’s viewer pane use the command below:
+
+radiant::radiant_viewer()
+To launch Radiant in an Rstudio Window use the command below:
+
+radiant::radiant_window()
+To easily update Radiant and the required packages, install the radiant.update package using:
+
+options(repos = c(RSM = "https://radiant-rstats.github.io/minicran", CRAN = "https://cloud.r-project.org"))
+install.packages("remotes")
+remotes::install_github("radiant-rstats/radiant.update", upgrade = "never")
+Then select Update radiant from the Addins menu in Rstudio or use the command below:
+
+radiant.update::radiant.update()
+See the installing radiant page additional for details.
+Optional: You can also create a launcher on your Desktop to start Radiant by typing radiant::launcher() in the R(studio) console and pressing return. A file called radiant.bat (windows) or radiant.command (mac) will be created that you can double-click to start Radiant in your default browser. The launcher command will also create a file called update_radiant.bat (windows) or update_radiant.command (mac) that you can double-click to update Radiant to the latest release.
+When Radiant starts you will see data on diamond prices. To close the application click the icon in the navigation bar and then click Stop. The Radiant process will stop and the browser window will close (Chrome) or gray-out.
+
+
+Documentation
+
+Documentation and tutorials are available at https://radiant-rstats.github.io/docs/ and in the Radiant web interface (the icons on each page and the icon in the navigation bar).
+Individual Radiant packages also each have their own pkgdown sites:
+
+- http://radiant-rstats.github.io/radiant
+- http://radiant-rstats.github.io/radiant.data
+- http://radiant-rstats.github.io/radiant.design
+- http://radiant-rstats.github.io/radiant.basics
+- http://radiant-rstats.github.io/radiant.model
+- http://radiant-rstats.github.io/radiant.multivariate
+
+Want some help getting started? Watch the tutorials on the documentation site.
+
+
+Reporting issues
+
+Please use the GitHub issue tracker at github.com/radiant-rstats/radiant/issues if you have any problems using Radiant.
+
+
+Try Radiant online
+
+Not ready to install Radiant on your computer? Try it online at the link below:
+https://vnijs.shinyapps.io/radiant
+Do not upload sensitive data to this public server. The size of data upload has been restricted to 10MB for security reasons.
+
+
+Running Radiant on shinyapps.io
+
+To run your own instance of Radiant on shinyapps.io first install Radiant and its dependencies. Then clone the radiant repo and ensure you have the latest version of the Radiant packages installed by running radiant/inst/app/for.shinyapps.io.R. Finally, open radiant/inst/app/ui.R and deploy the application.
+
+
+Running Radiant on shiny-server
+
+You can also host Radiant using shiny-server. First, install radiant on the server using the command below:
+
+options(repos = c(RSM = "https://radiant-rstats.github.io/minicran", CRAN = "https://cloud.r-project.org"))
+install.packages("radiant")
+Then clone the radiant repo and point shiny-server to the inst/app/ directory. As a courtesy, please let me know if you intend to use Radiant on a server.
+When running Radiant on a server, by default, file uploads are limited to 10MB and R-code in Report > Rmd and Report > R will not be evaluated for security reasons. If you have sudo access to the server and have appropriate security in place you can change these settings by adding the following lines to .Rprofile for the shiny user on the server.
+
+
+
+Running Radiant in the cloud (e.g., AWS)
+
+To run radiant in the cloud you can use the customized Docker container. See https://github.com/radiant-rstats/docker for details
+
+
+Saving and loading state
+
+To save your analyses save the state of the app to a file by clicking on the icon in the navbar and then on Save radiant state file (see also the Data > Manage tab). You can open this state file at a later time or on another computer to continue where you left off. You can also share the file with others that may want to replicate your analyses. As an example, load the state file radiant-example.state.rda by clicking on the icon in the navbar and then on Load radiant state file. Go to Data > View and Data > Visualize to see some of the settings from the previous “state” of the app. There is also a report in Report > Rmd that was created using the Radiant interface. The html file radiant-example.nb.html contains the output.
+A related feature in Radiant is that state is maintained if you accidentally navigate to another web page, close (and reopen) the browser, and/or hit refresh. Use Refresh in the menu in the navigation bar to return to a clean/new state.
+Loading and saving state also works with Rstudio. If you start Radiant from Rstudio and use > Stop to stop the app, lists called r_data, r_info, and r_state will be put into Rstudio’s global workspace. If you start radiant again using radiant::radiant() it will use these lists to restore state. Also, if you load a state file directly into Rstudio it will be used when you start Radiant to recreate a previous state.
+Technical note: Loading state works as follows in Radiant: When an input is initialized in a Shiny app you set a default value in the call to, for example, numericInput. In Radiant, when a state file has been loaded and an input is initialized it looks to see if there is a value for an input of that name in a list called r_state. If there is, this value is used. The r_state list is created when saving state using reactiveValuesToList(input). An example of a call to numericInput is given below where the state_init function from radiant.R is used to check if a value from r_state can be used.
+
+numericInput("sm_comp_value", "Comparison value:", state_init("sm_comp_value", 0))
+
+
+Source code
+
+The source code for the radiant application is available on GitHub at https://github.com/radiant-rstats. radiant.data, offers tools to load, save, view, visualize, summarize, combine, and transform data. radiant.design builds on radiant.data and adds tools for experimental design, sampling, and sample size calculation. radiant.basics covers the basics of statistical analysis (e.g., comparing means and proportions, cross-tabs, correlation, etc.) and includes a probability calculator. radiant.model covers model estimation (e.g., logistic regression and neural networks), model evaluation (e.g., gains chart, profit curve, confusion matrix, etc.), and decision tools (e.g., decision analysis and simulation). Finally, radiant.multivariate includes tools to generate brand maps and conduct cluster, factor, and conjoint analysis.
+These tools are used in the Business Analytics, Quantitative Analysis, Research for Marketing Decisions, Applied Market Research, Consumer Behavior, Experiments in Firms, Pricing, Pricing Analytics, and Customer Analytics classes at the Rady School of Management (UCSD).
+
+
+Credits
+
+Radiant would not be possible without R and Shiny. I would like to thank Joe Cheng, Winston Chang, and Yihui Xie for answering questions, providing suggestions, and creating amazing tools for the R community. Other key components used in Radiant are ggplot2, dplyr, tidyr, magrittr, broom, shinyAce, shinyFiles, rmarkdown, and DT. For an overview of other packages that Radiant relies on please see the about page.
+
+
+License
+
+Radiant is licensed under the AGPLv3. As a summary, the AGPLv3 license requires, attribution, including copyright and license information in copies of the software, stating changes if the code is modified, and disclosure of all source code. Details are in the COPYING file.
+The documentation, images, and videos for the radiant.data package are licensed under the creative commons attribution and share-alike license CC-BY-SA. All other documentation and videos on this site, as well as the help files for radiant.design, radiant.basics, radiant.model, and radiant.multivariate, are licensed under the creative commons attribution, non-commercial, share-alike license CC-NC-SA.
+If you are interested in using any of the radiant packages please email me at radiant@rady.ucsd.edu
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/radiant.data/docs/link.svg b/radiant.data/docs/link.svg
new file mode 100644
index 0000000000000000000000000000000000000000..88ad82769b87f10725c57dca6fcf41b4bffe462c
--- /dev/null
+++ b/radiant.data/docs/link.svg
@@ -0,0 +1,12 @@
+
+
+
diff --git a/radiant.data/docs/news/index.html b/radiant.data/docs/news/index.html
new file mode 100644
index 0000000000000000000000000000000000000000..82cfa04db83273ec8c0e4369d9df25cb8517e3c8
--- /dev/null
+++ b/radiant.data/docs/news/index.html
@@ -0,0 +1,727 @@
+
+Changelog • radiant.data
+
+
+
+
+
+
+
+
+
+
+ Changelog
+ Source: NEWS.md
+
+
+
+
+radiant.data 1.5.12023-01-09
+- Added features in the UI to facilitate persistent filters for filtered, sorted, and sliced data
+- Improvements to screenshot feature:
+
- Navigation bar is omitted and the image is adjusted to the length of the UI.
+- html2canvas.js is now included so users can take screenshot when offline
+
+- Added a convenience function
add_description to add a description attribute to a data.frame in markdown format
+- Line graphs treated more similarly to bar-graphs:
+
- Can have a binary factor variable on the y-axis
+- Y-variable only line are now also possible
+
+- Removed all references to
aes_string which is being deprecated in ggplot soon
+- Improved cleanup after Radiant UI is closed
+
+
+
+radiant.data 1.4.6
+- gsub(“[-]”, ““, text) is no longer valid in R 4.2.0 and above. Non-asci symbols will now be escaped using stringi::stri_trans_general when needed
+
+
+radiant.data 1.4.52022-09-08
+- Add scrolling for dropdown menus that might extend past the edge of the screen
+- Addressed warning messages about Font Awesome icons not existing
+- gsub(“[-]”, ““, text) is no longer valid in R 4.2.0 and above. Non-asci symbols will now be escaped using stringi when needed
+
+
+radiant.data 1.4.42022-07-19
+- Added option to create screenshots of settings on a page. Approach is inspired by the snapper package by @yonicd
+- Added contact request for users on Radiant startup
+- Fix issue with R_ZIPCMD when 7zip is on the path but not being recognized by R
+
+
+radiant.data 1.4.22022-05-25
+- Use
all for is.null and is.na if object length can be greater than 1 as required in R 4.2.0
+
+
+radiant.data 1.4.12021-11-12
+- Setup to allow formatting of the shiny interface with bootstrap 4
+- Addressing
is_empty function clash with rlang
+
+- Upgrading
shiny dependency to 1.6.0 and fixing project text alignment issue (@cpsievert, https://github.com/radiant-rstats/radiant.data/pull/28)
+
+
+radiant.data 1.3.122020-11-27
+- Fixes related to breaking changes in
magrittr
+
+- Fixes related to changes in
readr argument names
+- Fix to launch radiant in a “windows”
+
+
+radiant.data 1.3.102020-08-07
+- Add Google Drive to the default set of directories to explore if available
+- Add back functionality to convert a column to type
ts in Data > Transform now that this is again supported by dplyr 1.0.1
+
+
+radiant.data 1.3.92020-06-16
+- Fix for using the
date function from the lubridate package in a filter
+- Removed functionality to convert a column to type
ts as this is not supported by dplyr 1.0.0 and vctrs 0.3.1
+- Updated documentation using https://github.com/r-lib/roxygen2/pull/1109
+
+
+
+radiant.data 1.3.6
+- Updated styling for formatting for modals (e.g., help pages) that will also allow improved sizing of the (shinyFiles) file browser
+- Fix for
\r line-endings in Report > Rmd on Windows. Issue was most likely to occur when copy-and-pasting text from PDF into Report > Rmd.
+
+
+
+radiant.data 1.3.3
+- Function to calculate “mode”
+- Fix for “spread” in Data > Transform with column name includes “.”
+
+
+radiant.data 1.3.1
+- If radiant is not opened from an Rstudio project, use the working directory at launch as the base directory for the application
+
+
+radiant.data 1.3.0
+- Updated styling of Notebook and HTML reports (cosmo + zenburn)
+- Documentation updates to link to new video tutorials
+- Use
patchwork for grouping multiple plots together
+- Apply
refactor to any type in the Data > Transform UI
+- Fix for
weighted.sd when missing values differ for x and weights
+- Avoid resetting the “Column header” to its default value in Data > Explore when other settings are changed.
+
+
+radiant.data 1.2.3
+- Fix for Data > Transform > Spread when no variables are selected
+- Set
debounce to 0 for all shinyAce editors
+
+
+radiant.data 1.2.2
+- Use
zenburn for code highlighting in Notebook and HTML report from Report > Rmd
+
+- Clean up “sf_volumes” from the when radiant is stopped
+
+
+radiant.data 1.2.0
+- Update action buttons that initiate a calculation when one or more relevant inputs are changed. For example, when a model should be re-estimated because the set of explanatory variables was changed by the user, a spinning “refresh” icon will be shown
+
+
+radiant.data 1.1.8
+- Changed default
quantile algorithm used in the xtile function from number 2 to 7. See the help for stats::quantile for details
+- Added
me and meprop functions to calculate the margin of error for a mean and a proportion. Functions are accessible from Data > Pivot and Data > Explore
+
+
+
+radiant.data 1.1.6
+- Improvements for wrapping generated code to Report > Rmd or Report > R
+
+-
+Data > Transform > Training now uses the
randomizr package to allow blocking variables when creating a training variables.
+
+
+radiant.data 1.1.3
+- Guard against using Data > Transform > Reorder/remove levels with too many levels (i.e., > 100)
+- Guard against using Data > Transform > Reorder/remove variables with too many variables (i.e., > 100)
+- Fix for DT table callbacks when shiny 1.4 hits CRAN (see https://github.com/rstudio/DT/issues/146#issuecomment-534319155)
+- Tables from Data > Pivot and Data > Explore now have
nr set to Inf by default (i.e., show all rows). The user can change this to the number of desired rows to show (e.g., select 3 rows in a sorted table)
+- Fix for example numbering in the help file for Data > Transform
+
+- Numerous small code changes to support enhanced auto-completion, tooltips, and annotations in shinyAce 0.4.1
+
+
+radiant.data 1.0.62019-08-22
+- Fix for
Data > Transform > Change type
+
+- Option to
fix_names to lower case
+- Keyboard shortcut (Enter) to load remove csv and rds files
+- Use a shinyAce input to generate data descriptions
+- Allow custom initial dataset list
+- Fix for latex formulas in Report > Rmd on Windows
+- Updated requirements for markdown and Rmarkdown
+- Fix for
radiant.init.data with shiny-server
+- Improvements to setup to allow access to server-side files by adding options to .Rprofile:
+
- Add
options(radiant.report = TRUE) to allow report generation in Report > Rmd and Report > R
+
+- Add
options(radiant.shinyFiles = TRUE) to allow server-side access to files
+- List specific directories you want to use with radiant using, for example,
options(radiant.sf_volumes = c(Git = "/home/jovyan/git"))
+
+
+
+
+radiant.data 1.0.02019-07-18
+- Support for series of class
ts (e.g., Data > Transform > Change type > Time series)
+- Require shinyAce 0.4.0
+- Vertical jitter set to 0 by default
+
+
+radiant.data 0.9.9.0
+- Added option to save Report > Rmd as a powerpoint file using
Rmarkdown
+
+- Removed dependency on
summarytools due to breaking changes
+- Fix for interaction (
iterm) and non-linear term (qterm) creation if character strings rather than integers are passed to the function
+- Remove specific symbols from reports in Report > Rmd to avoid issues when generating HTML or PDF documents
+- Keyboard shortcuts, i.e., CTRL-O and CTRL-S (CMD-O and CMD-S on macOS) to open and save data files in the Data > Manage tab
+- Various fixes to address breaking changes in dplyr 0.8.0
+- Added
radiant_ prefix to all attributes, except description, to avoid conflicts with other packages (e.g., vars in dplyr)
+
+
+radiant.data 0.9.8.6
+- Use
stringi::stri_trans_general to replace special symbols in Rmarkdown that may cause problems
+- Add empty line before and after code chunks when saving reports to Rmarkdown
+- Use
rio to load sav, dta, or sas7bdat files through the read files button in Report > Rmd and Report > R.
+- Create a
qscatter plot similar to the function of the same name in Stata
+- New radiant icon
+- Fix for setting where both
xlim and ylim are set in visualize function
+- Use an expandable
shinyAce input for the R-code log in Data > Transform
+
+
+
+radiant.data 0.9.8.0
+- Added an “autosave” options. Use
options(radiant.autosave = c(10, 180)); radiant::radiant() to auto-save the application state to the ~/.radiant.session folder every 10 minutes for the next 180 minutes. This can be useful if radiant is being used during an exam, for example.
+- Emergency backups are now saved to
~/.radiant.session/r_some_id.state.rda. The files should be automatically loaded when needed but can also be loaded as a regular radiant state file
+- Replace option to load an
.rda from from a URL in Data > Manage to load .rds files instead
+- Ensure variable and dataset names are valid for R (i.e., no spaces or symbols), “fixing” the input as needed
+- Fix to visualize now
ggplot::labs no longer accepts a list as input
+- Add option to generate square and cubed terms for use in linear and logistic regression in
radiant.model
+
+- Fix for error when trying to save invalid predictions in
radiant.model. This action now generates a pop-up in the browser interface
+- Add a specified description to a data.frame immediately on
register
+
+- Option to pass additional arguments to
shiny::runApp when starting radiant such as the port to use. For example, radiant.data::radiant.data(“https://github.com/radiant-rstats/docs/raw/gh-pages/examples/demo-dvd-rnd.state.rda”, port = 8080)
+- Option for automatic cleanup of deprecated code in both Report > Rmd and Report > R
+
+- Avoid attempt to fix deprecated code in Report > Rmd if
pred_data = ""
+
+- Fix for download icon linked to downloading of a state file after upgrade to shiny 1.2
+- Update documentation for Data > Combine
+
+- Fix for
format_df when the data.frame contains missing values. This fix is relevant for several summary functions run in Report > Rmd or Report > R
+
+- Fix for directory set when using
Knit report in Report > Rmd and Report > R without an Rstudio project. Will now correctly default to the working directory used in R(studio)
+- Added option to change
smooth setting for histograms with a density plot
+- Similar to
pmin and pmax, pfun et al. calculate summary statistics elementwise across multiple vectors
+- Add
Desktop as a default directory to show in the shinyFiles file browser
+- Load a state file on startup by providing a (relative) file path or a url. For example, radiant.data::radiant.data(“https://github.com/radiant-rstats/docs/raw/gh-pages/examples/demo-dvd-rnd.state.rda”) or radiant.data::radiant.data(“assignment.state.rda”)
+- Update example report in Report > Rmd
+
+- Add
deregister function to remove data in radiant from memory and the datasets dropdown list
+- Fix for invalid column names if used in
Data > Pivot
+
+
+
+radiant.data 0.9.7.0
+- Use
summarytools to generate summary information for datasets in Data > Manage
+
+- Show modal with warning about non-writable working directory when saving reports in Report > Rmd or Report > R
+
+- Apply
radiant.data::fix_names to files loaded into radiant to ensure valid R-object names
+- Use the content of the
Store filtered data as input to name the csv download in Data > View
+
+- Add “txt” as a recognized file type for
Read files in Report > Rmd and Report > R
+
+- Allow multiple
lines or loess curves based on a selected color variable for scatter plots in Data > Visualize
+
+- Indicate that a plot in Data > Visualize should be updated when plot labels are changed
+- Fix for #81 when variables used in Data > Pivot contain dots
+- Fix for
radiant.project_dir when no Rstudio project is used which could cause incorrect relative paths to be used
+- Fix code formatting for Report > Rmd when arguments include a list (e.g., ggplot labels)
+- On Linux use a modal to show code in Report > Rmd and Report > R when reporting is set to “manual”
+- Use
is_double to ensure dates are not treated as numeric variables in Data > View
+
+- Make sort and filter state of tables in Data > Explore and Data > Pivot available in Report > Rmd
+- Fix names for data sets loaded using the
Read files button in Report > Rmd or Report > R
+- Cleanup environment after closing app
+- Fix column names with spaces, etc. when reading csv files
+- Additional styling and labeling options for Data > Visualize are now available in the browser interface
+- Fix for code generation related to DT filters
+
+
+radiant.data 0.9.6.14
+
+Major changes
+- Using
shinyFiles to provide convenient access to data located on a server
+- Avoid
XQuartz requirement
+
+
+Minor changes
+- Load
data(...) into the current environment rather than defaulting only to the global environment
+-
+
file.rename failed using docker on windows when saving a report. Using file.copy instead
+- Fix for
sf_volumes used to set the root directories to load and save files
+- Set default locale to “en_US.UTF-8” when using shiny-server unless
Sys.getlocale(category = "LC_ALL") what set to something other than “C”
+- Modal shown if and Rmd (R) file is not available when using “To Rstudio (Rmd)” in Report > Rmd or “To Rstudio (R)” in Report > R
+
+- Track progress loading (state) files
+- Fix for
radiant.sf_volumes used for the shinyFiles file browser
+- Improvements for sending code from Radiant to Rstudio
+- Better support for paths when using radiant on a server (i.e., revert to home directory using
radiant.data::find_home())
+- Revert from
svg to png for plots in _Report > Rmd_ and _Report > R_.svg` scatter plots with many point get to big for practical use on servers that have to transfer images to a local browser
+- Removed dependency on
methods package
+
+
+
+radiant.data 0.9.5.3
+- Fix smart comma’s in data descriptions
+- Search and replace
desc(n) in reports and replace by desc(n_obs)
+
+- Revert to storing the r_data environment as a list on stop to avoid reference problems (@josh1400)
+- Fix for plot type in Data > Pivot in older state files (@josh1400)
+- Used all declared imports (CRAN)
+
+
+radiant.data 0.9.5.0
+- Fix for
radiant.data::explore when variable names contain an underscore
+- Fix for
find_gdrive when drive is not being synced
+- Fixes in Report > Rmd and Report > R to accommodate for pandoc > 2
+
+
+radiant.data 0.9.4.6
+- Don’t update a reactive binding for an object if the binding already exists. See issue https://github.com/rstudio/shiny/issues/2065
+
+- Fix to accommodate changes in
deparse in R 3.5
+- Fix for saving data in Data > Manage and generating the relevant R-code
+
+
+
+
+radiant.data 0.9.3.3
+
+Major changes
+- When using radiant with Rstudio Viewer or in an Rstudio Window, loading and saving data through Data > Manage generates R-code the user can add to Report > Rmd or Report > R. Clicking the
Show R-code checkbox displays the R-code used to load or save the current dataset
+- Various changes to the code to accommodate the use of
shiny::makeReactiveBinding. The advantage is that the code generated for Report > Rmd and Report > R will no longer have to use a list (r_data) to store and access data. This means that code generated and used in the Radiant browser interface will be directly usable without the browser interface as well
+- Removed
loadr, saver, load_csv, loadcsv_url, loadrds_url, and make_funs functions as they are no longer needed
+- Deprecated
mean_rm, median_rm, min_rm, max_rm,sd_rm,var_rm, and sum_rm functions as they are no longer needed
+
+
+Minor changes
+- Added
load_clip and save_clip to load and save data to the clipboard on Windows and macOS
+- Improved auto completion in Report > Rmd and Report > R
+
+- Maintain, store, and clean the settings of the interactive table in Data > View
+
+- Address closing Rstudio Window issue (https://github.com/rstudio/shiny/issues/2033)
+
+
+
+radiant.data 0.9.2.3
+
+Major changes
+-
+Report > Rmd and Report > R will now be evaluated the
r_data environment. This means that the return value from ls() will be much cleaner
+
+
+Minor changes
+- Add option to load files with extension .rdata or .tsv using
loadr which add that data to the Datasets dropdown
+-
+
visualize will default to a scatter plot if xvar and yvar are specified but no plot type is provided in the function call
+- Improvements to
read_files function to interactively generate R-code (or Rmarkdown code-chunks) to read files in various format (e.g., SQLite, rds, csv, xlsx, css, jpg, etc.). Supports relative paths and uses find_dropbox() and find_gdrive() when applicable
+
+
+
+radiant.data 0.9.2.2
+
+Minor changes
+- Require
shinyAce 0.3.0
+- Export
read_files function to interactively generate R-code or Rmarkdown code-chunks to read files in various format (e.g., SQLite, rds, csv, xlsx, css, jpg, etc.). Supports relative paths and uses find_dropbox() and find_gdrive() when applicable
+
+
+
+
+CHANGES IN radiant.data 0.9.0.22
+
+Bug fixes
+- Fix for #43 where scatter plot was not shown for a dataset with less than 1,000 rows
+- Fix for Report > Rmd and Report > R when R-code or Rmarkdown is being pulled from the Rstudio editor
+
+
+
+
+
+radiant.data 0.9.0.16
+
+Minor changes
+- Changed license for help files and images for radiant.data to CC-BY-SA
+
+
+
+
+radiant.data 0.9.0.15
+
+
+
+
+radiant.data 0.9.0.13
+
+Minor changes
+- Apply
fixMS to replace curly quotes, em dash, etc. when using Data > Transform > Create
+
+- Option to set number of decimals to show in Data > View
+
+- Improved number formatting in interactive tables in Data > View, Data > Pivot, and Data > Explore
+
+- Option to include an interactive view of a dataset in Report > Rmd. By default, the number of rows is set to 100 as, most likely, the user will not want to embed a large dataset in save HTML report
+-
+Data > Transform will leave variables selected, unless switching to
Create or Spread
+
+- Switch focus to editor in Report > Rmd and Report > R when no other input has focus
+
+
+
+
+radiant.data 0.9.0.7
+
+Minor changes
+- Allow response variables with NA values in Model > Logistic regression and other classification models
+- Support logicals in code generation from Data > View
+
+- Track window size using
input$get_screen_width
+
+- Focus on editor when switching to Report > Rmd or Report > R so generated code is shown immediately and the user can navigate and type in the editor without having to click first
+- Add information about the first level when plotting a bar chart with a categorical variable on the Y-axis (e.g., mean(buyer {yes}))
+
+
+Bug fixes
+- Cleanup now also occurs when the stop button is used in Rstudio to close the app
+- Fix to include
DiagrammeR based plots in Rmarkdown reports
+- Fix in
read_files for SQLite data names
+- De-activate spell check auto correction in
selectizeInput in Rstudio Viewer shiny #1916
+
+- Fix to allow selecting and copying text output from Report > Rmd and Report > R
+
+- Remove “fancy” quotes from filters
+- Known issue: The Rstudio viewer may not always close the viewer window when trying to stop the application with the
Stop link in the navbar. As a work-around, use Rstudio’s stop buttons instead.
+
+
+
+radiant.data 0.9.0.0
+
+Major changes
+- If Rstudio project is used Report > Rmd and Report > R will use the project directory as base. This allows users to use relative paths and making it easier to share (reproducible) code
+- Specify options in .Rprofile for upload memory limit and running Report > Rmd on server
+-
+
find_project function based on rstudioapi
+
+-
+Report > Rmd Read button to generate code to load various types of data (e.g., rda, rds, xls, yaml, feather)
+-
+Report > Rmd Read button to generate code to load various types of files in report (e.g., jpg, png, md, Rmd, R). If Radiant was started from an Rstudio project, the file paths used will be relative to the project root. Paths to files synced to local Dropbox or Google Drive folder will use the
find_dropbox and find_gdrive functions to enhances reproducibility.
+-
+Report > Rmd Load Report button can be used to load Rmarkdown file in the editor. It will also extract the source code from Notebook and HTML files with embedded Rmarkdown
+-
+Report > Rmd will read Rmd directly from Rstudio when “To Rstudio (Rmd)” is selected. This will make it possible to use Rstudio Server Pro’s Share project option for realtime collaboration in Radiant
+- Long lines of code generated for Report > Rmd will be wrapped to enhance readability
+-
+Report > R is now equivalent to Report > Rmd but in R-code format
+-
+Report > Rmd option to view Editor, Preview, or Both
+- Show Rstudio project information in navbar if available
+
+
+
+
+radiant.data 0.8.7.8
+
+Minor changes
+- Added preview options to Data > Manage based on https://github.com/radiant-rstats/radiant/issues/30
+
+- Add selected dataset name as default table download name in Data > View, Data > Pivot, and Data > Explore
+
+- Use “stack” as the default for histograms and frequency charts in Data > Visualize
+
+- Cleanup
Stop & Report option in navbar
+- Upgraded tidyr dependency to 0.7
+- Upgraded dplyr dependency to 0.7.1
+
+
+
+
+radiant.data 0.8.6.0
+
+Minor changes
+- Export
ggplotly from plotly for interactive plots in Report > Rmd
+
+- Export
subplot from plotly for grids of interactive plots in Report > Rmd
+
+- Set default
res = 96 for renderPlot and dpi = 96 for knitr::opts_chunk
+
+- Add
fillcol, linecol, and pointcol to visualize to set plot colors when no fill or color variable has been selected
+- Reverse legend ordering in Data > Visualize when axes are flipped using
coor_flip()
+
+- Added functions to choose.files and choose.dir. Uses JavaScript on Mac, utils::choose.files and utils::choose.dir on Windows, and reverts to file.choose on Linux
+- Added
find_gdrive to determine the path to a user’s local Google Drive folder if available
+-
+
fixMs for encoding in reports on Windows
+
+
+
+
+radiant.data 0.8.1.0
+
+Minor changes
+- Specify the maximum number of rows to load for a csv and csv (url) file through Data > Manage
+
+- Support for loading and saving feather files, including specifying the maximum number of rows to load through Data > Manage
+
+- Added author and year arguments to help modals in inst/app/radiant.R (thanks @kmezhoud)
+- Added size argument for scatter plots to create bubble charts (thanks @andrewsali)
+- Example and CSS formatting for tables in Report > Rmd
+
+- Added
seed argument to make_train
+
+- Added
prop, sdprop, etc. for working with proportions
+- Set
ylim in visualize for multiple plots
+- Show progress indicator when saving reports from Report > Rmd
+
+-
+
copy_attr convenience function
+-
+
refactor function to keep only a subset of levels in a factor and recode the remaining (and first) level to, for example, other
+-
+
register function to add a (transformed) dataset to the dataset dropdown
+- Remember name of state files loaded and suggest that name when re-saving the state
+- Show dataset name in output if dataframe passed directly to analysis function
+- R-notebooks are now the default option for output saved from Report > Rmd and Report > R
+
+- Improved documentation on how to customize plots in Report > Rmd
+
+- Keyboard short-cut to put code into Report > Rmd (ALT-enter)
+
+
+Bug fixes
+- When clicking the
rename button, without changing the name, the dataset was set to NULL (thanks @kmezhoud, https://github.com/radiant-rstats/radiant/issues/5)
+- Replace ext with .ext in
mutate_each function call
+- Variance estimation in Data > Explore would cause an error with unit cell-frequencies (thanks @kmezhoud, https://github.com/radiant-rstats/radiant/issues/6)
+- Fix for as_integer when factor levels are characters
+- Fix for integer conversion in explore
+- Remove \r and special characters from strings in r_data and r_state
+- Fix sorting in Report > Rmd for tables created using Data > Pivot and Data > Explore when column headers contain symbols or spaces (thanks @4kammer)
+- Set
error = TRUE for rmarkdown for consistency with knitr as used in Report > Rmd
+
+- Correctly handle decimal indicators when loading csv files in Data > Manage
+
+- Don’t overwrite a dataset to combine if combine generates an error when user sets the the name of the combined data to that of an already selected dataset
+- When multiple variables were selected, data were not correctly summarized in Data > Transform
+- Add (function) label to bar plot when x-variable is an integer
+- Maintain order of variables in Data > Visualize when using “color”, “fill”, “comby”, or “combx”
+- Avoid warning when switching datasets in Data > Transform and variables being summarized do not exists in the new dataset
+- which.pmax produced a list but needed to be integer
+- To customized predictions in radiant.model indexr must be able to customize the prediction dataframe
+- describe now correctly resets the working directory on exit
+- removed all calls to summarise_each and mutate_each from dplyr
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/radiant.data/docs/pkgdown.css b/radiant.data/docs/pkgdown.css
new file mode 100644
index 0000000000000000000000000000000000000000..80ea5b838a4d8f24ae023bbf3582a8079aadbf66
--- /dev/null
+++ b/radiant.data/docs/pkgdown.css
@@ -0,0 +1,384 @@
+/* Sticky footer */
+
+/**
+ * Basic idea: https://philipwalton.github.io/solved-by-flexbox/demos/sticky-footer/
+ * Details: https://github.com/philipwalton/solved-by-flexbox/blob/master/assets/css/components/site.css
+ *
+ * .Site -> body > .container
+ * .Site-content -> body > .container .row
+ * .footer -> footer
+ *
+ * Key idea seems to be to ensure that .container and __all its parents__
+ * have height set to 100%
+ *
+ */
+
+html, body {
+ height: 100%;
+}
+
+body {
+ position: relative;
+}
+
+body > .container {
+ display: flex;
+ height: 100%;
+ flex-direction: column;
+}
+
+body > .container .row {
+ flex: 1 0 auto;
+}
+
+footer {
+ margin-top: 45px;
+ padding: 35px 0 36px;
+ border-top: 1px solid #e5e5e5;
+ color: #666;
+ display: flex;
+ flex-shrink: 0;
+}
+footer p {
+ margin-bottom: 0;
+}
+footer div {
+ flex: 1;
+}
+footer .pkgdown {
+ text-align: right;
+}
+footer p {
+ margin-bottom: 0;
+}
+
+img.icon {
+ float: right;
+}
+
+/* Ensure in-page images don't run outside their container */
+.contents img {
+ max-width: 100%;
+ height: auto;
+}
+
+/* Fix bug in bootstrap (only seen in firefox) */
+summary {
+ display: list-item;
+}
+
+/* Typographic tweaking ---------------------------------*/
+
+.contents .page-header {
+ margin-top: calc(-60px + 1em);
+}
+
+dd {
+ margin-left: 3em;
+}
+
+/* Section anchors ---------------------------------*/
+
+a.anchor {
+ display: none;
+ margin-left: 5px;
+ width: 20px;
+ height: 20px;
+
+ background-image: url(./link.svg);
+ background-repeat: no-repeat;
+ background-size: 20px 20px;
+ background-position: center center;
+}
+
+h1:hover .anchor,
+h2:hover .anchor,
+h3:hover .anchor,
+h4:hover .anchor,
+h5:hover .anchor,
+h6:hover .anchor {
+ display: inline-block;
+}
+
+/* Fixes for fixed navbar --------------------------*/
+
+.contents h1, .contents h2, .contents h3, .contents h4 {
+ padding-top: 60px;
+ margin-top: -40px;
+}
+
+/* Navbar submenu --------------------------*/
+
+.dropdown-submenu {
+ position: relative;
+}
+
+.dropdown-submenu>.dropdown-menu {
+ top: 0;
+ left: 100%;
+ margin-top: -6px;
+ margin-left: -1px;
+ border-radius: 0 6px 6px 6px;
+}
+
+.dropdown-submenu:hover>.dropdown-menu {
+ display: block;
+}
+
+.dropdown-submenu>a:after {
+ display: block;
+ content: " ";
+ float: right;
+ width: 0;
+ height: 0;
+ border-color: transparent;
+ border-style: solid;
+ border-width: 5px 0 5px 5px;
+ border-left-color: #cccccc;
+ margin-top: 5px;
+ margin-right: -10px;
+}
+
+.dropdown-submenu:hover>a:after {
+ border-left-color: #ffffff;
+}
+
+.dropdown-submenu.pull-left {
+ float: none;
+}
+
+.dropdown-submenu.pull-left>.dropdown-menu {
+ left: -100%;
+ margin-left: 10px;
+ border-radius: 6px 0 6px 6px;
+}
+
+/* Sidebar --------------------------*/
+
+#pkgdown-sidebar {
+ margin-top: 30px;
+ position: -webkit-sticky;
+ position: sticky;
+ top: 70px;
+}
+
+#pkgdown-sidebar h2 {
+ font-size: 1.5em;
+ margin-top: 1em;
+}
+
+#pkgdown-sidebar h2:first-child {
+ margin-top: 0;
+}
+
+#pkgdown-sidebar .list-unstyled li {
+ margin-bottom: 0.5em;
+}
+
+/* bootstrap-toc tweaks ------------------------------------------------------*/
+
+/* All levels of nav */
+
+nav[data-toggle='toc'] .nav > li > a {
+ padding: 4px 20px 4px 6px;
+ font-size: 1.5rem;
+ font-weight: 400;
+ color: inherit;
+}
+
+nav[data-toggle='toc'] .nav > li > a:hover,
+nav[data-toggle='toc'] .nav > li > a:focus {
+ padding-left: 5px;
+ color: inherit;
+ border-left: 1px solid #878787;
+}
+
+nav[data-toggle='toc'] .nav > .active > a,
+nav[data-toggle='toc'] .nav > .active:hover > a,
+nav[data-toggle='toc'] .nav > .active:focus > a {
+ padding-left: 5px;
+ font-size: 1.5rem;
+ font-weight: 400;
+ color: inherit;
+ border-left: 2px solid #878787;
+}
+
+/* Nav: second level (shown on .active) */
+
+nav[data-toggle='toc'] .nav .nav {
+ display: none; /* Hide by default, but at >768px, show it */
+ padding-bottom: 10px;
+}
+
+nav[data-toggle='toc'] .nav .nav > li > a {
+ padding-left: 16px;
+ font-size: 1.35rem;
+}
+
+nav[data-toggle='toc'] .nav .nav > li > a:hover,
+nav[data-toggle='toc'] .nav .nav > li > a:focus {
+ padding-left: 15px;
+}
+
+nav[data-toggle='toc'] .nav .nav > .active > a,
+nav[data-toggle='toc'] .nav .nav > .active:hover > a,
+nav[data-toggle='toc'] .nav .nav > .active:focus > a {
+ padding-left: 15px;
+ font-weight: 500;
+ font-size: 1.35rem;
+}
+
+/* orcid ------------------------------------------------------------------- */
+
+.orcid {
+ font-size: 16px;
+ color: #A6CE39;
+ /* margins are required by official ORCID trademark and display guidelines */
+ margin-left:4px;
+ margin-right:4px;
+ vertical-align: middle;
+}
+
+/* Reference index & topics ----------------------------------------------- */
+
+.ref-index th {font-weight: normal;}
+
+.ref-index td {vertical-align: top; min-width: 100px}
+.ref-index .icon {width: 40px;}
+.ref-index .alias {width: 40%;}
+.ref-index-icons .alias {width: calc(40% - 40px);}
+.ref-index .title {width: 60%;}
+
+.ref-arguments th {text-align: right; padding-right: 10px;}
+.ref-arguments th, .ref-arguments td {vertical-align: top; min-width: 100px}
+.ref-arguments .name {width: 20%;}
+.ref-arguments .desc {width: 80%;}
+
+/* Nice scrolling for wide elements --------------------------------------- */
+
+table {
+ display: block;
+ overflow: auto;
+}
+
+/* Syntax highlighting ---------------------------------------------------- */
+
+pre, code, pre code {
+ background-color: #f8f8f8;
+ color: #333;
+}
+pre, pre code {
+ white-space: pre-wrap;
+ word-break: break-all;
+ overflow-wrap: break-word;
+}
+
+pre {
+ border: 1px solid #eee;
+}
+
+pre .img, pre .r-plt {
+ margin: 5px 0;
+}
+
+pre .img img, pre .r-plt img {
+ background-color: #fff;
+}
+
+code a, pre a {
+ color: #375f84;
+}
+
+a.sourceLine:hover {
+ text-decoration: none;
+}
+
+.fl {color: #1514b5;}
+.fu {color: #000000;} /* function */
+.ch,.st {color: #036a07;} /* string */
+.kw {color: #264D66;} /* keyword */
+.co {color: #888888;} /* comment */
+
+.error {font-weight: bolder;}
+.warning {font-weight: bolder;}
+
+/* Clipboard --------------------------*/
+
+.hasCopyButton {
+ position: relative;
+}
+
+.btn-copy-ex {
+ position: absolute;
+ right: 0;
+ top: 0;
+ visibility: hidden;
+}
+
+.hasCopyButton:hover button.btn-copy-ex {
+ visibility: visible;
+}
+
+/* headroom.js ------------------------ */
+
+.headroom {
+ will-change: transform;
+ transition: transform 200ms linear;
+}
+.headroom--pinned {
+ transform: translateY(0%);
+}
+.headroom--unpinned {
+ transform: translateY(-100%);
+}
+
+/* mark.js ----------------------------*/
+
+mark {
+ background-color: rgba(255, 255, 51, 0.5);
+ border-bottom: 2px solid rgba(255, 153, 51, 0.3);
+ padding: 1px;
+}
+
+/* vertical spacing after htmlwidgets */
+.html-widget {
+ margin-bottom: 10px;
+}
+
+/* fontawesome ------------------------ */
+
+.fab {
+ font-family: "Font Awesome 5 Brands" !important;
+}
+
+/* don't display links in code chunks when printing */
+/* source: https://stackoverflow.com/a/10781533 */
+@media print {
+ code a:link:after, code a:visited:after {
+ content: "";
+ }
+}
+
+/* Section anchors ---------------------------------
+ Added in pandoc 2.11: https://github.com/jgm/pandoc-templates/commit/9904bf71
+*/
+
+div.csl-bib-body { }
+div.csl-entry {
+ clear: both;
+}
+.hanging-indent div.csl-entry {
+ margin-left:2em;
+ text-indent:-2em;
+}
+div.csl-left-margin {
+ min-width:2em;
+ float:left;
+}
+div.csl-right-inline {
+ margin-left:2em;
+ padding-left:1em;
+}
+div.csl-indent {
+ margin-left: 2em;
+}
diff --git a/radiant.data/docs/pkgdown.js b/radiant.data/docs/pkgdown.js
new file mode 100644
index 0000000000000000000000000000000000000000..6f0eee40bc3c69061d33fbed1bc9644f47c0eb8a
--- /dev/null
+++ b/radiant.data/docs/pkgdown.js
@@ -0,0 +1,108 @@
+/* http://gregfranko.com/blog/jquery-best-practices/ */
+(function($) {
+ $(function() {
+
+ $('.navbar-fixed-top').headroom();
+
+ $('body').css('padding-top', $('.navbar').height() + 10);
+ $(window).resize(function(){
+ $('body').css('padding-top', $('.navbar').height() + 10);
+ });
+
+ $('[data-toggle="tooltip"]').tooltip();
+
+ var cur_path = paths(location.pathname);
+ var links = $("#navbar ul li a");
+ var max_length = -1;
+ var pos = -1;
+ for (var i = 0; i < links.length; i++) {
+ if (links[i].getAttribute("href") === "#")
+ continue;
+ // Ignore external links
+ if (links[i].host !== location.host)
+ continue;
+
+ var nav_path = paths(links[i].pathname);
+
+ var length = prefix_length(nav_path, cur_path);
+ if (length > max_length) {
+ max_length = length;
+ pos = i;
+ }
+ }
+
+ // Add class to parent
Radiant is an open-source platform-independent browser-based interface for business analytics in R. The application is based on the Shiny package and can be run locally or on a server. Radiant was developed by Vincent Nijs. Please use the issue tracker on GitHub to suggest enhancements or report problems: https://github.com/radiant-rstats/radiant.data/issues. For other questions and comments please use radiant@rady.ucsd.edu.
+Key features +
+-
+
- Explore: Quickly and easily summarize, visualize, and analyze your data +
- Cross-platform: It runs in a browser on Windows, Mac, and Linux +
- Reproducible: Recreate results and share work with others as a state file or an Rmarkdown report +
- Programming: Integrate Radiant’s analysis functions with your own R-code +
- Context: Data and examples focus on business applications +
Playlists +
+There are two youtube playlists with video tutorials. The first provides a general introduction to key features in Radiant. The second covers topics relevant in a course on business analytics (i.e., Probability, Decision Analysis, Hypothesis Testing, Linear Regression, and Simulation).
+ +Explore +
+Radiant is interactive. Results update immediately when inputs are changed (i.e., no separate dialog boxes) and/or when a button is pressed (e.g., Estimate in Model > Estimate > Logistic regression (GLM)). This facilitates rapid exploration and understanding of the data.
Cross-platform +
+Radiant works on Windows, Mac, or Linux. It can run without an Internet connection and no data will leave your computer. You can also run the app as a web application on a server.
+Reproducible +
+To conduct high-quality analysis, simply saving output is not enough. You need the ability to reproduce results for the same data and/or when new data become available. Moreover, others may want to review your analysis and results. Save and load the state of the application to continue your work at a later time or on another computer. Share state files with others and create reproducible reports using Rmarkdown. See also the section on Saving and loading state below
If you are using Radiant on a server you can even share the URL (include the SSUID) with others so they can see what you are working on. Thanks for this feature go to Joe Cheng.
+Programming +
+Although Radiant’s web-interface can handle quite a few data and analysis tasks, you may prefer to write your own R-code. Radiant provides a bridge to programming in R(studio) by exporting the functions used for analysis (i.e., you can conduct your analysis using the Radiant web-interface or by calling Radiant’s functions directly from R-code). For more information about programming with Radiant see the programming page on the documentation site.
+How to install Radiant +
+ +In Rstudio you can start and update Radiant through the Addins menu at the top of the screen. To install the latest version of Radiant for Windows or Mac, with complete documentation for off-line access, open R(studio) and copy-and-paste the command below:
+options(repos = c(RSM = "https://radiant-rstats.github.io/minicran", CRAN = "https://cloud.r-project.org"))
+install.packages("radiant")Once all packages are installed, select Start radiant from the Addins menu in Rstudio or use the command below to launch the app:
+radiant::radiant()To launch Radiant in Rstudio’s viewer pane use the command below:
+
+radiant::radiant_viewer()To launch Radiant in an Rstudio Window use the command below:
+
+radiant::radiant_window()To easily update Radiant and the required packages, install the radiant.update package using:
+options(repos = c(RSM = "https://radiant-rstats.github.io/minicran", CRAN = "https://cloud.r-project.org"))
+install.packages("remotes")
+remotes::install_github("radiant-rstats/radiant.update", upgrade = "never")Then select Update radiant from the Addins menu in Rstudio or use the command below:
+radiant.update::radiant.update()See the installing radiant page additional for details.
+Optional: You can also create a launcher on your Desktop to start Radiant by typing radiant::launcher() in the R(studio) console and pressing return. A file called radiant.bat (windows) or radiant.command (mac) will be created that you can double-click to start Radiant in your default browser. The launcher command will also create a file called update_radiant.bat (windows) or update_radiant.command (mac) that you can double-click to update Radiant to the latest release.
When Radiant starts you will see data on diamond prices. To close the application click the icon in the navigation bar and then click Stop. The Radiant process will stop and the browser window will close (Chrome) or gray-out.
Documentation +
+Documentation and tutorials are available at https://radiant-rstats.github.io/docs/ and in the Radiant web interface (the icons on each page and the icon in the navigation bar).
+Individual Radiant packages also each have their own pkgdown sites:
+-
+
- http://radiant-rstats.github.io/radiant +
- http://radiant-rstats.github.io/radiant.data +
- http://radiant-rstats.github.io/radiant.design +
- http://radiant-rstats.github.io/radiant.basics +
- http://radiant-rstats.github.io/radiant.model +
- http://radiant-rstats.github.io/radiant.multivariate +
Want some help getting started? Watch the tutorials on the documentation site.
+Reporting issues +
+Please use the GitHub issue tracker at github.com/radiant-rstats/radiant/issues if you have any problems using Radiant.
+Try Radiant online +
+Not ready to install Radiant on your computer? Try it online at the link below:
+https://vnijs.shinyapps.io/radiant
+Do not upload sensitive data to this public server. The size of data upload has been restricted to 10MB for security reasons.
+Running Radiant on shinyapps.io +
+To run your own instance of Radiant on shinyapps.io first install Radiant and its dependencies. Then clone the radiant repo and ensure you have the latest version of the Radiant packages installed by running radiant/inst/app/for.shinyapps.io.R. Finally, open radiant/inst/app/ui.R and deploy the application.
Running Radiant on shiny-server +
+You can also host Radiant using shiny-server. First, install radiant on the server using the command below:
+
+options(repos = c(RSM = "https://radiant-rstats.github.io/minicran", CRAN = "https://cloud.r-project.org"))
+install.packages("radiant")Then clone the radiant repo and point shiny-server to the inst/app/ directory. As a courtesy, please let me know if you intend to use Radiant on a server.
When running Radiant on a server, by default, file uploads are limited to 10MB and R-code in Report > Rmd and Report > R will not be evaluated for security reasons. If you have sudo access to the server and have appropriate security in place you can change these settings by adding the following lines to .Rprofile for the shiny user on the server.
Running Radiant in the cloud (e.g., AWS) +
+To run radiant in the cloud you can use the customized Docker container. See https://github.com/radiant-rstats/docker for details
+Saving and loading state +
+To save your analyses save the state of the app to a file by clicking on the icon in the navbar and then on Save radiant state file (see also the Data > Manage tab). You can open this state file at a later time or on another computer to continue where you left off. You can also share the file with others that may want to replicate your analyses. As an example, load the state file radiant-example.state.rda by clicking on the icon in the navbar and then on Load radiant state file. Go to Data > View and Data > Visualize to see some of the settings from the previous “state” of the app. There is also a report in Report > Rmd that was created using the Radiant interface. The html file radiant-example.nb.html contains the output.
A related feature in Radiant is that state is maintained if you accidentally navigate to another web page, close (and reopen) the browser, and/or hit refresh. Use Refresh in the menu in the navigation bar to return to a clean/new state.
Loading and saving state also works with Rstudio. If you start Radiant from Rstudio and use > Stop to stop the app, lists called r_data, r_info, and r_state will be put into Rstudio’s global workspace. If you start radiant again using radiant::radiant() it will use these lists to restore state. Also, if you load a state file directly into Rstudio it will be used when you start Radiant to recreate a previous state.
Technical note: Loading state works as follows in Radiant: When an input is initialized in a Shiny app you set a default value in the call to, for example, numericInput. In Radiant, when a state file has been loaded and an input is initialized it looks to see if there is a value for an input of that name in a list called r_state. If there is, this value is used. The r_state list is created when saving state using reactiveValuesToList(input). An example of a call to numericInput is given below where the state_init function from radiant.R is used to check if a value from r_state can be used.
+numericInput("sm_comp_value", "Comparison value:", state_init("sm_comp_value", 0))Source code +
+The source code for the radiant application is available on GitHub at https://github.com/radiant-rstats. radiant.data, offers tools to load, save, view, visualize, summarize, combine, and transform data. radiant.design builds on radiant.data and adds tools for experimental design, sampling, and sample size calculation. radiant.basics covers the basics of statistical analysis (e.g., comparing means and proportions, cross-tabs, correlation, etc.) and includes a probability calculator. radiant.model covers model estimation (e.g., logistic regression and neural networks), model evaluation (e.g., gains chart, profit curve, confusion matrix, etc.), and decision tools (e.g., decision analysis and simulation). Finally, radiant.multivariate includes tools to generate brand maps and conduct cluster, factor, and conjoint analysis.
These tools are used in the Business Analytics, Quantitative Analysis, Research for Marketing Decisions, Applied Market Research, Consumer Behavior, Experiments in Firms, Pricing, Pricing Analytics, and Customer Analytics classes at the Rady School of Management (UCSD).
+Credits +
+Radiant would not be possible without R and Shiny. I would like to thank Joe Cheng, Winston Chang, and Yihui Xie for answering questions, providing suggestions, and creating amazing tools for the R community. Other key components used in Radiant are ggplot2, dplyr, tidyr, magrittr, broom, shinyAce, shinyFiles, rmarkdown, and DT. For an overview of other packages that Radiant relies on please see the about page.
+License +
+Radiant is licensed under the AGPLv3. As a summary, the AGPLv3 license requires, attribution, including copyright and license information in copies of the software, stating changes if the code is modified, and disclosure of all source code. Details are in the COPYING file.
+The documentation, images, and videos for the radiant.data package are licensed under the creative commons attribution and share-alike license CC-BY-SA. All other documentation and videos on this site, as well as the help files for radiant.design, radiant.basics, radiant.model, and radiant.multivariate, are licensed under the creative commons attribution, non-commercial, share-alike license CC-NC-SA.
If you are interested in using any of the radiant packages please email me at radiant@rady.ucsd.edu
+ +Changelog
+ Source:NEWS.md
+ radiant.data 1.5.12023-01-09
+- Added features in the UI to facilitate persistent filters for filtered, sorted, and sliced data +
- Improvements to screenshot feature:
+
- Navigation bar is omitted and the image is adjusted to the length of the UI. +
- html2canvas.js is now included so users can take screenshot when offline +
+ - Added a convenience function
add_descriptionto add a description attribute to a data.frame in markdown format
+ - Line graphs treated more similarly to bar-graphs:
+
- Can have a binary factor variable on the y-axis +
- Y-variable only line are now also possible +
+ - Removed all references to
aes_stringwhich is being deprecated in ggplot soon
+ - Improved cleanup after Radiant UI is closed +
radiant.data 1.4.6
+- gsub(“[-]”, ““, text) is no longer valid in R 4.2.0 and above. Non-asci symbols will now be escaped using stringi::stri_trans_general when needed +
radiant.data 1.4.52022-09-08
+- Add scrolling for dropdown menus that might extend past the edge of the screen +
- Addressed warning messages about Font Awesome icons not existing +
- gsub(“[-]”, ““, text) is no longer valid in R 4.2.0 and above. Non-asci symbols will now be escaped using stringi when needed +
radiant.data 1.4.42022-07-19
+- Added option to create screenshots of settings on a page. Approach is inspired by the snapper package by @yonicd +
- Added contact request for users on Radiant startup +
- Fix issue with R_ZIPCMD when 7zip is on the path but not being recognized by R +
radiant.data 1.4.22022-05-25
+- Use
allforis.nullandis.naif object length can be greater than 1 as required in R 4.2.0
+
radiant.data 1.4.12021-11-12
+- Setup to allow formatting of the shiny interface with bootstrap 4 +
- Addressing
is_emptyfunction clash withrlang+
+ - Upgrading
shinydependency to 1.6.0 and fixing project text alignment issue (@cpsievert, https://github.com/radiant-rstats/radiant.data/pull/28)
+
radiant.data 1.3.122020-11-27
+- Fixes related to breaking changes in
magrittr+
+ - Fixes related to changes in
readrargument names
+ - Fix to launch radiant in a “windows” +
radiant.data 1.3.102020-08-07
+- Add Google Drive to the default set of directories to explore if available +
- Add back functionality to convert a column to type
tsin Data > Transform now that this is again supported by dplyr 1.0.1
+
radiant.data 1.3.92020-06-16
+- Fix for using the
datefunction from the lubridate package in a filter
+ - Removed functionality to convert a column to type
tsas this is not supported by dplyr 1.0.0 and vctrs 0.3.1
+ - Updated documentation using https://github.com/r-lib/roxygen2/pull/1109 + +
radiant.data 1.3.6
+- Updated styling for formatting for modals (e.g., help pages) that will also allow improved sizing of the (shinyFiles) file browser +
- Fix for
\rline-endings in Report > Rmd on Windows. Issue was most likely to occur when copy-and-pasting text from PDF into Report > Rmd.
+
radiant.data 1.3.3
+- Function to calculate “mode” +
- Fix for “spread” in Data > Transform with column name includes “.” +
radiant.data 1.3.1
+- If radiant is not opened from an Rstudio project, use the working directory at launch as the base directory for the application +
radiant.data 1.3.0
+- Updated styling of Notebook and HTML reports (cosmo + zenburn) +
- Documentation updates to link to new video tutorials +
- Use
patchworkfor grouping multiple plots together
+ - Apply
refactorto any type in the Data > Transform UI
+ - Fix for
weighted.sdwhen missing values differ forxand weights
+ - Avoid resetting the “Column header” to its default value in Data > Explore when other settings are changed. +
radiant.data 1.2.3
+- Fix for Data > Transform > Spread when no variables are selected +
- Set
debounceto 0 for all shinyAce editors
+
radiant.data 1.2.2
+- Use
zenburnfor code highlighting in Notebook and HTML report from Report > Rmd +
+ - Clean up “sf_volumes” from the when radiant is stopped +
radiant.data 1.2.0
+- Update action buttons that initiate a calculation when one or more relevant inputs are changed. For example, when a model should be re-estimated because the set of explanatory variables was changed by the user, a spinning “refresh” icon will be shown +
radiant.data 1.1.8
+- Changed default
quantilealgorithm used in thextilefunction from number 2 to 7. See the help forstats::quantilefor details
+ - Added
meandmepropfunctions to calculate the margin of error for a mean and a proportion. Functions are accessible from Data > Pivot and Data > Explore +
+
radiant.data 1.1.6
+- Improvements for wrapping generated code to Report > Rmd or Report > R + +
-
+Data > Transform > Training now uses the
randomizrpackage to allow blocking variables when creating a training variables.
+
radiant.data 1.1.3
+- Guard against using Data > Transform > Reorder/remove levels with too many levels (i.e., > 100) +
- Guard against using Data > Transform > Reorder/remove variables with too many variables (i.e., > 100) +
- Fix for DT table callbacks when shiny 1.4 hits CRAN (see https://github.com/rstudio/DT/issues/146#issuecomment-534319155) +
- Tables from Data > Pivot and Data > Explore now have
nrset toInfby default (i.e., show all rows). The user can change this to the number of desired rows to show (e.g., select 3 rows in a sorted table)
+ - Fix for example numbering in the help file for Data > Transform + +
- Numerous small code changes to support enhanced auto-completion, tooltips, and annotations in shinyAce 0.4.1 +
radiant.data 1.0.62019-08-22
+- Fix for
Data > Transform > Change type+
+ - Option to
fix_namesto lower case
+ - Keyboard shortcut (Enter) to load remove csv and rds files +
- Use a shinyAce input to generate data descriptions +
- Allow custom initial dataset list +
- Fix for latex formulas in Report > Rmd on Windows +
- Updated requirements for markdown and Rmarkdown +
- Fix for
radiant.init.datawith shiny-server
+ - Improvements to setup to allow access to server-side files by adding options to .Rprofile:
+
- Add
options(radiant.report = TRUE)to allow report generation in Report > Rmd and Report > R +
+ - Add
options(radiant.shinyFiles = TRUE)to allow server-side access to files
+ - List specific directories you want to use with radiant using, for example,
options(radiant.sf_volumes = c(Git = "/home/jovyan/git"))+
+
+ - Add
radiant.data 1.0.02019-07-18
+- Support for series of class
ts(e.g., Data > Transform > Change type > Time series)
+ - Require shinyAce 0.4.0 +
- Vertical jitter set to 0 by default +
radiant.data 0.9.9.0
+- Added option to save Report > Rmd as a powerpoint file using
Rmarkdown+
+ - Removed dependency on
summarytoolsdue to breaking changes
+ - Fix for interaction (
iterm) and non-linear term (qterm) creation if character strings rather than integers are passed to the function
+ - Remove specific symbols from reports in Report > Rmd to avoid issues when generating HTML or PDF documents +
- Keyboard shortcuts, i.e., CTRL-O and CTRL-S (CMD-O and CMD-S on macOS) to open and save data files in the Data > Manage tab +
- Various fixes to address breaking changes in dplyr 0.8.0 +
- Added
radiant_prefix to all attributes, exceptdescription, to avoid conflicts with other packages (e.g.,varsin dplyr)
+
radiant.data 0.9.8.6
+- Use
stringi::stri_trans_generalto replace special symbols in Rmarkdown that may cause problems
+ - Add empty line before and after code chunks when saving reports to Rmarkdown +
- Use
rioto loadsav,dta, orsas7bdatfiles through theread filesbutton in Report > Rmd and Report > R.
+ - Create a
qscatterplot similar to the function of the same name in Stata
+ - New radiant icon +
- Fix for setting where both
xlimandylimare set invisualizefunction
+ - Use an expandable
shinyAceinput for the R-code log in Data > Transform +
+
radiant.data 0.9.8.0
+- Added an “autosave” options. Use
options(radiant.autosave = c(10, 180)); radiant::radiant()to auto-save the application state to the~/.radiant.sessionfolder every 10 minutes for the next 180 minutes. This can be useful if radiant is being used during an exam, for example.
+ - Emergency backups are now saved to
~/.radiant.session/r_some_id.state.rda. The files should be automatically loaded when needed but can also be loaded as a regular radiant state file
+ - Replace option to load an
.rdafrom from a URL in Data > Manage to load.rdsfiles instead
+ - Ensure variable and dataset names are valid for R (i.e., no spaces or symbols), “fixing” the input as needed +
- Fix to visualize now
ggplot::labsno longer accepts a list as input
+ - Add option to generate square and cubed terms for use in linear and logistic regression in
radiant.model+
+ - Fix for error when trying to save invalid predictions in
radiant.model. This action now generates a pop-up in the browser interface
+ - Add a specified description to a data.frame immediately on
register+
+ - Option to pass additional arguments to
shiny::runAppwhen starting radiant such as the port to use. For example, radiant.data::radiant.data(“https://github.com/radiant-rstats/docs/raw/gh-pages/examples/demo-dvd-rnd.state.rda”, port = 8080)
+ - Option for automatic cleanup of deprecated code in both Report > Rmd and Report > R + +
- Avoid attempt to fix deprecated code in Report > Rmd if
pred_data = ""+
+ - Fix for download icon linked to downloading of a state file after upgrade to shiny 1.2 +
- Update documentation for Data > Combine + +
- Fix for
format_dfwhen the data.frame contains missing values. This fix is relevant for severalsummaryfunctions run in Report > Rmd or Report > R +
+ - Fix for directory set when using
Knit reportin Report > Rmd and Report > R without an Rstudio project. Will now correctly default to the working directory used in R(studio)
+ - Added option to change
smoothsetting for histograms with a density plot
+ - Similar to
pminandpmax,pfunet al. calculate summary statistics elementwise across multiple vectors
+ - Add
Desktopas a default directory to show in theshinyFilesfile browser
+ - Load a state file on startup by providing a (relative) file path or a url. For example, radiant.data::radiant.data(“https://github.com/radiant-rstats/docs/raw/gh-pages/examples/demo-dvd-rnd.state.rda”) or radiant.data::radiant.data(“assignment.state.rda”) +
- Update example report in Report > Rmd + +
- Add
deregisterfunction to remove data in radiant from memory and thedatasetsdropdown list
+ - Fix for invalid column names if used in
Data > Pivot+
+
radiant.data 0.9.7.0
+- Use
summarytoolsto generate summary information for datasets in Data > Manage +
+ - Show modal with warning about non-writable working directory when saving reports in Report > Rmd or Report > R + +
- Apply
radiant.data::fix_namesto files loaded into radiant to ensure valid R-object names
+ - Use the content of the
Store filtered data asinput to name the csv download in Data > View +
+ - Add “txt” as a recognized file type for
Read filesin Report > Rmd and Report > R +
+ - Allow multiple
linesorloesscurves based on a selectedcolorvariable for scatter plots in Data > Visualize +
+ - Indicate that a plot in Data > Visualize should be updated when plot labels are changed +
- Fix for #81 when variables used in Data > Pivot contain dots +
- Fix for
radiant.project_dirwhen no Rstudio project is used which could cause incorrect relative paths to be used
+ - Fix code formatting for Report > Rmd when arguments include a list (e.g., ggplot labels) +
- On Linux use a modal to show code in Report > Rmd and Report > R when reporting is set to “manual” +
- Use
is_doubleto ensure dates are not treated as numeric variables in Data > View +
+ - Make sort and filter state of tables in Data > Explore and Data > Pivot available in Report > Rmd +
- Fix names for data sets loaded using the
Read filesbutton in Report > Rmd or Report > R
+ - Cleanup environment after closing app +
- Fix column names with spaces, etc. when reading csv files +
- Additional styling and labeling options for Data > Visualize are now available in the browser interface +
- Fix for code generation related to DT filters +
radiant.data 0.9.6.14
+Major changes
+- Using
shinyFilesto provide convenient access to data located on a server
+ - Avoid
XQuartzrequirement
+
Minor changes
+- Load
data(...)into the current environment rather than defaulting only to the global environment
+ -
+
file.renamefailed using docker on windows when saving a report. Usingfile.copyinstead
+ - Fix for
sf_volumesused to set the root directories to load and save files
+ - Set default locale to “en_US.UTF-8” when using shiny-server unless
Sys.getlocale(category = "LC_ALL")what set to something other than “C”
+ - Modal shown if and Rmd (R) file is not available when using “To Rstudio (Rmd)” in Report > Rmd or “To Rstudio (R)” in Report > R + +
- Track progress loading (state) files +
- Fix for
radiant.sf_volumesused for theshinyFilesfile browser
+ - Improvements for sending code from Radiant to Rstudio +
- Better support for paths when using radiant on a server (i.e., revert to home directory using
radiant.data::find_home())
+ - Revert from
svgtopngfor plots in_Report > Rmd_ and _Report > R_.svg` scatter plots with many point get to big for practical use on servers that have to transfer images to a local browser
+ - Removed dependency on
methodspackage
+
radiant.data 0.9.5.3
+- Fix smart comma’s in data descriptions +
- Search and replace
desc(n)in reports and replace bydesc(n_obs)+
+ - Revert to storing the r_data environment as a list on stop to avoid reference problems (@josh1400) +
- Fix for plot type in Data > Pivot in older state files (@josh1400) +
- Used all declared imports (CRAN) +
radiant.data 0.9.5.0
+- Fix for
radiant.data::explorewhen variable names contain an underscore
+ - Fix for
find_gdrivewhen drive is not being synced
+ - Fixes in Report > Rmd and Report > R to accommodate for pandoc > 2 +
radiant.data 0.9.4.6
+- Don’t update a reactive binding for an object if the binding already exists. See issue https://github.com/rstudio/shiny/issues/2065 + +
- Fix to accommodate changes in
deparsein R 3.5
+ - Fix for saving data in Data > Manage and generating the relevant R-code +
radiant.data 0.9.3.3
+Major changes
+- When using radiant with Rstudio Viewer or in an Rstudio Window, loading and saving data through Data > Manage generates R-code the user can add to Report > Rmd or Report > R. Clicking the
Show R-codecheckbox displays the R-code used to load or save the current dataset
+ - Various changes to the code to accommodate the use of
shiny::makeReactiveBinding. The advantage is that the code generated for Report > Rmd and Report > R will no longer have to use a list (r_data) to store and access data. This means that code generated and used in the Radiant browser interface will be directly usable without the browser interface as well
+ - Removed
loadr,saver,load_csv,loadcsv_url,loadrds_url, andmake_funsfunctions as they are no longer needed
+ - Deprecated
mean_rm,median_rm,min_rm,max_rm,sd_rm,var_rm, andsum_rmfunctions as they are no longer needed
+
Minor changes
+- Added
load_clipandsave_clipto load and save data to the clipboard on Windows and macOS
+ - Improved auto completion in Report > Rmd and Report > R + +
- Maintain, store, and clean the settings of the interactive table in Data > View + +
- Address closing Rstudio Window issue (https://github.com/rstudio/shiny/issues/2033) +
radiant.data 0.9.2.3
+Major changes
+-
+Report > Rmd and Report > R will now be evaluated the
r_dataenvironment. This means that the return value fromls()will be much cleaner
+
Minor changes
+- Add option to load files with extension .rdata or .tsv using
loadrwhich add that data to the Datasets dropdown
+ -
+
visualizewill default to a scatter plot ifxvarandyvarare specified but no plottypeis provided in the function call
+ - Improvements to
read_filesfunction to interactively generate R-code (or Rmarkdown code-chunks) to read files in various format (e.g., SQLite, rds, csv, xlsx, css, jpg, etc.). Supports relative paths and usesfind_dropbox()andfind_gdrive()when applicable
+
radiant.data 0.9.2.2
+Minor changes
+- Require
shinyAce0.3.0
+ - Export
read_filesfunction to interactively generate R-code or Rmarkdown code-chunks to read files in various format (e.g., SQLite, rds, csv, xlsx, css, jpg, etc.). Supports relative paths and usesfind_dropbox()andfind_gdrive()when applicable
+
CHANGES IN radiant.data 0.9.0.22
+Bug fixes
+- Fix for #43 where scatter plot was not shown for a dataset with less than 1,000 rows +
- Fix for Report > Rmd and Report > R when R-code or Rmarkdown is being pulled from the Rstudio editor +
radiant.data 0.9.0.16
+Minor changes
+- Changed license for help files and images for radiant.data to CC-BY-SA + +
radiant.data 0.9.0.15
+ + +radiant.data 0.9.0.13
+Minor changes
+- Apply
fixMSto replace curly quotes, em dash, etc. when using Data > Transform > Create +
+ - Option to set number of decimals to show in Data > View + +
- Improved number formatting in interactive tables in Data > View, Data > Pivot, and Data > Explore + +
- Option to include an interactive view of a dataset in Report > Rmd. By default, the number of rows is set to 100 as, most likely, the user will not want to embed a large dataset in save HTML report +
-
+Data > Transform will leave variables selected, unless switching to
CreateorSpread+
+ - Switch focus to editor in Report > Rmd and Report > R when no other input has focus +
radiant.data 0.9.0.7
+Minor changes
+- Allow response variables with NA values in Model > Logistic regression and other classification models +
- Support logicals in code generation from Data > View + +
- Track window size using
input$get_screen_width+
+ - Focus on editor when switching to Report > Rmd or Report > R so generated code is shown immediately and the user can navigate and type in the editor without having to click first +
- Add information about the first level when plotting a bar chart with a categorical variable on the Y-axis (e.g., mean(buyer {yes})) +
Bug fixes
+- Cleanup now also occurs when the stop button is used in Rstudio to close the app +
- Fix to include
DiagrammeRbased plots in Rmarkdown reports
+ - Fix in
read_filesfor SQLite data names
+ - De-activate spell check auto correction in
selectizeInputin Rstudio Viewer shiny #1916 +
+ - Fix to allow selecting and copying text output from Report > Rmd and Report > R + +
- Remove “fancy” quotes from filters +
- Known issue: The Rstudio viewer may not always close the viewer window when trying to stop the application with the
Stoplink in the navbar. As a work-around, use Rstudio’s stop buttons instead.
+
radiant.data 0.9.0.0
+Major changes
+- If Rstudio project is used Report > Rmd and Report > R will use the project directory as base. This allows users to use relative paths and making it easier to share (reproducible) code +
- Specify options in .Rprofile for upload memory limit and running Report > Rmd on server +
-
+
find_projectfunction based onrstudioapi+
+ - +Report > Rmd Read button to generate code to load various types of data (e.g., rda, rds, xls, yaml, feather) +
-
+Report > Rmd Read button to generate code to load various types of files in report (e.g., jpg, png, md, Rmd, R). If Radiant was started from an Rstudio project, the file paths used will be relative to the project root. Paths to files synced to local Dropbox or Google Drive folder will use the
find_dropboxandfind_gdrivefunctions to enhances reproducibility.
+ - +Report > Rmd Load Report button can be used to load Rmarkdown file in the editor. It will also extract the source code from Notebook and HTML files with embedded Rmarkdown +
- +Report > Rmd will read Rmd directly from Rstudio when “To Rstudio (Rmd)” is selected. This will make it possible to use Rstudio Server Pro’s Share project option for realtime collaboration in Radiant +
- Long lines of code generated for Report > Rmd will be wrapped to enhance readability +
- +Report > R is now equivalent to Report > Rmd but in R-code format +
- +Report > Rmd option to view Editor, Preview, or Both +
- Show Rstudio project information in navbar if available +
radiant.data 0.8.7.8
+Minor changes
+- Added preview options to Data > Manage based on https://github.com/radiant-rstats/radiant/issues/30 + +
- Add selected dataset name as default table download name in Data > View, Data > Pivot, and Data > Explore + +
- Use “stack” as the default for histograms and frequency charts in Data > Visualize + +
- Cleanup
Stop & Reportoption in navbar
+ - Upgraded tidyr dependency to 0.7 +
- Upgraded dplyr dependency to 0.7.1 +
radiant.data 0.8.6.0
+Minor changes
+- Export
ggplotlyfromplotlyfor interactive plots in Report > Rmd +
+ - Export
subplotfromplotlyfor grids of interactive plots in Report > Rmd +
+ - Set default
res = 96forrenderPlotanddpi = 96forknitr::opts_chunk+
+ - Add
fillcol,linecol, andpointcoltovisualizeto set plot colors when nofillorcolorvariable has been selected
+ - Reverse legend ordering in Data > Visualize when axes are flipped using
coor_flip()+
+ - Added functions to choose.files and choose.dir. Uses JavaScript on Mac, utils::choose.files and utils::choose.dir on Windows, and reverts to file.choose on Linux +
- Added
find_gdriveto determine the path to a user’s local Google Drive folder if available
+ -
+
fixMsfor encoding in reports on Windows
+
radiant.data 0.8.1.0
+Minor changes
+- Specify the maximum number of rows to load for a csv and csv (url) file through Data > Manage + +
- Support for loading and saving feather files, including specifying the maximum number of rows to load through Data > Manage + +
- Added author and year arguments to help modals in inst/app/radiant.R (thanks @kmezhoud) +
- Added size argument for scatter plots to create bubble charts (thanks @andrewsali) +
- Example and CSS formatting for tables in Report > Rmd + +
- Added
seedargument tomake_train+
+ - Added
prop,sdprop, etc. for working with proportions
+ - Set
yliminvisualizefor multiple plots
+ - Show progress indicator when saving reports from Report > Rmd + +
-
+
copy_attrconvenience function
+ -
+
refactorfunction to keep only a subset of levels in a factor and recode the remaining (and first) level to, for example, other
+ -
+
registerfunction to add a (transformed) dataset to the dataset dropdown
+ - Remember name of state files loaded and suggest that name when re-saving the state +
- Show dataset name in output if dataframe passed directly to analysis function +
- R-notebooks are now the default option for output saved from Report > Rmd and Report > R + +
- Improved documentation on how to customize plots in Report > Rmd + +
- Keyboard short-cut to put code into Report > Rmd (ALT-enter) +
Bug fixes
+- When clicking the
renamebutton, without changing the name, the dataset was set to NULL (thanks @kmezhoud, https://github.com/radiant-rstats/radiant/issues/5)
+ - Replace ext with .ext in
mutate_eachfunction call
+ - Variance estimation in Data > Explore would cause an error with unit cell-frequencies (thanks @kmezhoud, https://github.com/radiant-rstats/radiant/issues/6) +
- Fix for as_integer when factor levels are characters +
- Fix for integer conversion in explore +
- Remove \r and special characters from strings in r_data and r_state +
- Fix sorting in Report > Rmd for tables created using Data > Pivot and Data > Explore when column headers contain symbols or spaces (thanks @4kammer) +
- Set
error = TRUEfor rmarkdown for consistency with knitr as used in Report > Rmd +
+ - Correctly handle decimal indicators when loading csv files in Data > Manage + +
- Don’t overwrite a dataset to combine if combine generates an error when user sets the the name of the combined data to that of an already selected dataset +
- When multiple variables were selected, data were not correctly summarized in Data > Transform +
- Add (function) label to bar plot when x-variable is an integer +
- Maintain order of variables in Data > Visualize when using “color”, “fill”, “comby”, or “combx” +
- Avoid warning when switching datasets in Data > Transform and variables being summarized do not exists in the new dataset +
- which.pmax produced a list but needed to be integer +
- To customized predictions in radiant.model indexr must be able to customize the prediction dataframe +
- describe now correctly resets the working directory on exit +
- removed all calls to summarise_each and mutate_each from dplyr +
Convenience function to add a class
+add_class(x, cl)Arguments
+- x +
Object
+
+
+- cl +
Vector of class labels to add
+
+
Examples
+foo <- "some text" %>% add_class("text")
+foo <- "some text" %>% add_class(c("text", "another class"))
+Convenience function to add a markdown description to a data.frame
+ Source:R/radiant.R
+ add_description.RdConvenience function to add a markdown description to a data.frame
+add_description(df, md = "", path = "")Arguments
+- df +
A data.frame or tibble
+
+
+- md +
Data description in markdown format
+
+
+- path +
Path to a text file with the data description in markdown format
+
+
See also
+See also register
Examples
+if (interactive()) {
+ mt <- mtcars |> add_description(md = "# MTCARS\n\nThis data.frame contains information on ...")
+ describe(mt)
+}
+
+Arrange data with user-specified expression
+arrange_data(dataset, expr = NULL)Arguments
+- dataset +
Data frame to arrange
+
+
+- expr +
Expression to use arrange rows from the specified dataset
+
+
Value
+ + +Arranged data frame
+Details
+Arrange data, likely in combination with slicing
+Wrapper for as.character
+as_character(x)Arguments
+- x +
Input vector
+
+
Distance in kilometers or miles between two locations based on lat-long +Function based on http://www.movable-type.co.uk/scripts/latlong.html. Uses the haversine formula
+ Source:R/transform.R
+ as_distance.RdDistance in kilometers or miles between two locations based on lat-long +Function based on http://www.movable-type.co.uk/scripts/latlong.html. Uses the haversine formula
+as_distance(
+ lat1,
+ long1,
+ lat2,
+ long2,
+ unit = "km",
+ R = c(km = 6371, miles = 3959)[[unit]]
+)Arguments
+- lat1 +
Latitude of location 1
+
+
+- long1 +
Longitude of location 1
+
+
+- lat2 +
Latitude of location 2
+
+
+- long2 +
Longitude of location 2
+
+
+- unit +
Measure kilometers ("km", default) or miles ("miles")
+
+
+- R +
Radius of the earth
+
+
Value
+ + +Distance between two points
+Examples
+as_distance(32.8245525, -117.0951632, 40.7033127, -73.979681, unit = "km")
+#> [1] 3898.601
+as_distance(32.8245525, -117.0951632, 40.7033127, -73.979681, unit = "miles")
+#> [1] 2422.628
+
+Convert input in day-month-year format to date
+as_dmy(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Date variable of class Date
+Examples
+as_dmy("1-2-2014")
+#> [1] "2014-02-01"
+
+Convert input in day-month-year-hour-minute format to date-time
+ Source:R/transform.R
+ as_dmy_hm.RdConvert input in day-month-year-hour-minute format to date-time
+as_dmy_hm(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Date-time variable of class Date
+Examples
+as_mdy_hm("1-1-2014 12:15")
+#> [1] "2014-01-01 12:15:00 UTC"
+Convert input in day-month-year-hour-minute-second format to date-time
+ Source:R/transform.R
+ as_dmy_hms.RdConvert input in day-month-year-hour-minute-second format to date-time
+as_dmy_hms(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Date-time variable of class Date
+Examples
+as_mdy_hms("1-1-2014 12:15:01")
+#> [1] "2014-01-01 12:15:01 UTC"
+Wrapper for lubridate's as.duration function. Result converted to numeric
+ Source:R/transform.R
+ as_duration.RdWrapper for lubridate's as.duration function. Result converted to numeric
+as_duration(x)Arguments
+- x +
Time difference
+
+
Wrapper for factor with ordered = FALSE
+as_factor(x, ordered = FALSE)Arguments
+- x +
Input vector
+
+
+- ordered +
Order factor levels (TRUE, FALSE)
+
+
Convert input in hour-minute format to time
+as_hm(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Time variable of class Period
+Examples
+as_hm("12:45")
+#> [1] "12H 45M 0S"
+if (FALSE) {
+as_hm("12:45") %>% minute()
+}
+Convert input in hour-minute-second format to time
+as_hms(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Time variable of class Period
+Examples
+as_hms("12:45:00")
+#> [1] "12H 45M 0S"
+if (FALSE) {
+as_hms("12:45:00") %>% hour()
+as_hms("12:45:00") %>% second()
+}
+Convert variable to integer avoiding potential issues with factors
+ Source:R/transform.R
+ as_integer.RdConvert variable to integer avoiding potential issues with factors
+as_integer(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Integer
+Examples
+as_integer(rnorm(10))
+#> [1] 0 0 0 0 1 0 0 1 0 1
+as_integer(letters)
+#> [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25
+#> [26] 26
+as_integer(as.factor(5:10))
+#> [1] 5 6 7 8 9 10
+as.integer(as.factor(5:10))
+#> [1] 1 2 3 4 5 6
+as_integer(c("a", "b"))
+#> [1] 1 2
+as_integer(c("0", "1"))
+#> [1] 0 1
+as_integer(as.factor(c("0", "1")))
+#> [1] 0 1
+
+Convert input in month-day-year format to date
+as_mdy(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Date variable of class Date
+Details
+Use as.character if x is a factor
+Convert input in month-day-year-hour-minute format to date-time
+ Source:R/transform.R
+ as_mdy_hm.RdConvert input in month-day-year-hour-minute format to date-time
+as_mdy_hm(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Date-time variable of class Date
+Examples
+as_mdy_hm("1-1-2014 12:15")
+#> [1] "2014-01-01 12:15:00 UTC"
+Convert input in month-day-year-hour-minute-second format to date-time
+ Source:R/transform.R
+ as_mdy_hms.RdConvert input in month-day-year-hour-minute-second format to date-time
+as_mdy_hms(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Date-time variable of class Date
+Examples
+as_mdy_hms("1-1-2014 12:15:01")
+#> [1] "2014-01-01 12:15:01 UTC"
+Convert variable to numeric avoiding potential issues with factors
+ Source:R/transform.R
+ as_numeric.RdConvert variable to numeric avoiding potential issues with factors
+as_numeric(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Numeric
+Examples
+as_numeric(rnorm(10))
+#> [1] -1.66539524 0.84589239 -0.14666561 1.15801832 0.47822510 0.15957457
+#> [7] -0.42560129 -1.27584256 0.02811642 -0.40987108
+as_numeric(letters)
+#> [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25
+#> [26] 26
+as_numeric(as.factor(5:10))
+#> [1] 5 6 7 8 9 10
+as.numeric(as.factor(5:10))
+#> [1] 1 2 3 4 5 6
+as_numeric(c("a", "b"))
+#> [1] 1 2
+as_numeric(c("3", "4"))
+#> [1] 3 4
+as_numeric(as.factor(c("3", "4")))
+#> [1] 3 4
+
+Convert input in year-month-day format to date
+as_ymd(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Date variable of class Date
+Examples
+as_ymd("2013-1-1")
+#> [1] "2013-01-01"
+
+Convert input in year-month-day-hour-minute format to date-time
+ Source:R/transform.R
+ as_ymd_hm.RdConvert input in year-month-day-hour-minute format to date-time
+as_ymd_hm(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Date-time variable of class Date
+Examples
+as_ymd_hm("2014-1-1 12:15")
+#> [1] "2014-01-01 12:15:00 UTC"
+Convert input in year-month-day-hour-minute-second format to date-time
+ Source:R/transform.R
+ as_ymd_hms.RdConvert input in year-month-day-hour-minute-second format to date-time
+as_ymd_hms(x)Arguments
+- x +
Input variable
+
+
Value
+ + +Date-time variable of class Date
+Center
+center(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +If x is a numeric variable return x - mean(x)
+Choose a directory interactively
+choose_dir(...)Arguments
+- ... +
Arguments passed to utils::choose.dir on Windows
+
+
Value
+ + +Path to the directory selected by the user
+Details
+Open a file dialog to select a directory. Uses JavaScript on Mac, utils::choose.dir on Windows, and dirname(file.choose()) on Linux
+Examples
+if (FALSE) {
+choose_dir()
+}
+
+Choose files interactively
+choose_files(...)Arguments
+- ... +
Strings used to indicate which file types should be available for selection (e.g., "csv" or "pdf")
+
+
Value
+ + +Vector of paths to files selected by the user
+Details
+Open a file dialog. Uses JavaScript on Mac, utils::choose.files on Windows, and file.choose() on Linux
+Examples
+if (FALSE) {
+choose_files("pdf", "csv")
+}
+
+Labels for confidence intervals
+ci_label(alt = "two.sided", cl = 0.95, dec = 3)Arguments
+- alt +
Type of hypothesis ("two.sided","less","greater")
+
+
+- cl +
Confidence level
+
+
+- dec +
Number of decimals to show
+
+
Value
+ + +A character vector with labels for a confidence interval
+Examples
+ci_label("less", .95)
+#> [1] "0%" "95%"
+ci_label("two.sided", .95)
+#> [1] "2.5%" "97.5%"
+ci_label("greater", .9)
+#> [1] "10%" "100%"
+Values at confidence levels
+ci_perc(dat, alt = "two.sided", cl = 0.95)Arguments
+- dat +
Data
+
+
+- alt +
Type of hypothesis ("two.sided","less","greater")
+
+
+- cl +
Confidence level
+
+
Value
+ + +A vector with values at a confidence level
+Examples
+ci_perc(0:100, "less", .95)
+#> 5%
+#> 5
+ci_perc(0:100, "greater", .95)
+#> 95%
+#> 95
+ci_perc(0:100, "two.sided", .80)
+#> 10% 90%
+#> 10 90
+Combine datasets using dplyr's bind and join functions
+combine_data(
+ x,
+ y,
+ by = "",
+ add = "",
+ type = "inner_join",
+ data_filter = "",
+ arr = "",
+ rows = NULL,
+ envir = parent.frame(),
+ ...
+)Arguments
+- x +
Dataset
+
+
+- y +
Dataset to combine with x
+
+
+- by +
Variables used to combine `x` and `y`
+
+
+- add +
Variables to add from `y`
+
+
+- type +
The main bind and join types from the dplyr package are provided. inner_join returns all rows from x with matching values in y, and all columns from x and y. If there are multiple matches between x and y, all match combinations are returned. left_join returns all rows from x, and all columns from x and y. If there are multiple matches between x and y, all match combinations are returned. right_join is equivalent to a left join for datasets y and x. full_join combines two datasets, keeping rows and columns that appear in either. semi_join returns all rows from x with matching values in y, keeping just columns from x. A semi join differs from an inner join because an inner join will return one row of x for each matching row of y, whereas a semi join will never duplicate rows of x. anti_join returns all rows from x without matching values in y, keeping only columns from x. bind_rows and bind_cols are also included, as are intersect, union, and setdiff. See https://radiant-rstats.github.io/docs/data/combine.html for further details
+
+
+- data_filter +
Expression used to filter the dataset. This should be a string (e.g., "price > 10000")
+
+
+- arr +
Expression to arrange (sort) the data on (e.g., "color, desc(price)")
+
+
+- rows +
Rows to select from the specified dataset
+
+
+- envir +
Environment to extract data from
+
+
+- ... +
further arguments passed to or from other methods
+
+
Value
+ + +Combined dataset
+Details
+See https://radiant-rstats.github.io/docs/data/combine.html for an example in Radiant
+Examples
+avengers %>% combine_data(superheroes, type = "bind_cols")
+#> New names:
+#> • `name` -> `name...1`
+#> • `alignment` -> `alignment...2`
+#> • `gender` -> `gender...3`
+#> • `publisher` -> `publisher...4`
+#> • `name` -> `name...5`
+#> • `alignment` -> `alignment...6`
+#> • `gender` -> `gender...7`
+#> • `publisher` -> `publisher...8`
+#> # A tibble: 7 × 8
+#> name...1 alignment...2 gender...3 publisher...4 name...5 alignment...6
+#> <chr> <chr> <chr> <chr> <chr> <chr>
+#> 1 Thor good male Marvel Magneto bad
+#> 2 Iron Man good male Marvel Storm good
+#> 3 Hulk good male Marvel Mystique bad
+#> 4 Hawkeye good male Marvel Batman good
+#> 5 Black Widow good female Marvel Joker bad
+#> 6 Captain America good male Marvel Catwoman bad
+#> 7 Magneto bad male Marvel Hellboy good
+#> # ℹ 2 more variables: gender...7 <chr>, publisher...8 <chr>
+combine_data(avengers, superheroes, type = "bind_cols")
+#> New names:
+#> • `name` -> `name...1`
+#> • `alignment` -> `alignment...2`
+#> • `gender` -> `gender...3`
+#> • `publisher` -> `publisher...4`
+#> • `name` -> `name...5`
+#> • `alignment` -> `alignment...6`
+#> • `gender` -> `gender...7`
+#> • `publisher` -> `publisher...8`
+#> # A tibble: 7 × 8
+#> name...1 alignment...2 gender...3 publisher...4 name...5 alignment...6
+#> <chr> <chr> <chr> <chr> <chr> <chr>
+#> 1 Thor good male Marvel Magneto bad
+#> 2 Iron Man good male Marvel Storm good
+#> 3 Hulk good male Marvel Mystique bad
+#> 4 Hawkeye good male Marvel Batman good
+#> 5 Black Widow good female Marvel Joker bad
+#> 6 Captain America good male Marvel Catwoman bad
+#> 7 Magneto bad male Marvel Hellboy good
+#> # ℹ 2 more variables: gender...7 <chr>, publisher...8 <chr>
+avengers %>% combine_data(superheroes, type = "bind_rows")
+#> # A tibble: 14 × 4
+#> name alignment gender publisher
+#> <chr> <chr> <chr> <chr>
+#> 1 Thor good male Marvel
+#> 2 Iron Man good male Marvel
+#> 3 Hulk good male Marvel
+#> 4 Hawkeye good male Marvel
+#> 5 Black Widow good female Marvel
+#> 6 Captain America good male Marvel
+#> 7 Magneto bad male Marvel
+#> 8 Magneto bad male Marvel
+#> 9 Storm good female Marvel
+#> 10 Mystique bad female Marvel
+#> 11 Batman good male DC
+#> 12 Joker bad male DC
+#> 13 Catwoman bad female DC
+#> 14 Hellboy good male Dark Horse Comics
+avengers %>% combine_data(superheroes, add = "publisher", type = "bind_rows")
+#> # A tibble: 14 × 4
+#> name alignment gender publisher
+#> <chr> <chr> <chr> <chr>
+#> 1 Thor good male Marvel
+#> 2 Iron Man good male Marvel
+#> 3 Hulk good male Marvel
+#> 4 Hawkeye good male Marvel
+#> 5 Black Widow good female Marvel
+#> 6 Captain America good male Marvel
+#> 7 Magneto bad male Marvel
+#> 8 Magneto bad male Marvel
+#> 9 Storm good female Marvel
+#> 10 Mystique bad female Marvel
+#> 11 Batman good male DC
+#> 12 Joker bad male DC
+#> 13 Catwoman bad female DC
+#> 14 Hellboy good male Dark Horse Comics
+
+Source all package functions
+copy_all(.from)Arguments
+- .from +
The package to pull the function from
+
+
Details
+Equivalent of source with local=TRUE for all package functions. Adapted from functions by smbache, author of the import package. See https://github.com/rticulate/import/issues/4/ for a discussion. This function will be deprecated when (if) it is included in https://github.com/rticulate/import/
+Examples
+copy_all(radiant.data)
+Copy attributes from one object to another
+copy_attr(to, from, attr)Arguments
+- to +
Object to copy attributes to
+
+
+- from +
Object to copy attributes from
+
+
+- attr +
Vector of attributes. If missing all attributes will be copied
+
+
Source for package functions
+copy_from(.from, ...)Arguments
+- .from +
The package to pull the function from
+
+
+- ... +
Functions to pull
+
+
Details
+Equivalent of source with local=TRUE for package functions. Written by smbache, author of the import package. See https://github.com/rticulate/import/issues/4/ for a discussion. This function will be deprecated when (if) it is included in https://github.com/rticulate/import/
+Examples
+copy_from(radiant.data, get_data)
+Coefficient of variation
+cv(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Coefficient of variation
+Examples
+cv(runif(100))
+#> [1] 0.5724333
+
+Deregister a data.frame or list in Radiant
+deregister(
+ dataset,
+ shiny = shiny::getDefaultReactiveDomain(),
+ envir = r_data,
+ info = r_info
+)Arguments
+- dataset +
String containing the name of the data.frame to deregister
+
+
+- shiny +
Check if function is called from a shiny application
+
+
+- envir +
Environment to remove data from
+
+
+- info +
Reactive list with information about available data in radiant
+
+
Show dataset description
+describe(dataset, envir = parent.frame())Arguments
+- dataset +
Dataset with "description" attribute
+
+
+- envir +
Environment to extract data from
+
+
Details
+Show dataset description, if available, in html form in Rstudio viewer or the default browser. The description should be in markdown format, attached to a data.frame as an attribute with the name "description"
+Does a vector have non-zero variability?
+does_vary(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Logical. TRUE is there is variability
+Examples
+summarise_all(diamonds, does_vary) %>% as.logical()
+#> [1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
+
+Create an interactive table to view, search, sort, and filter data
+ Source:R/view.R
+ dtab.data.frame.RdCreate an interactive table to view, search, sort, and filter data
+# S3 method for data.frame
+dtab(
+ object,
+ vars = "",
+ filt = "",
+ arr = "",
+ rows = NULL,
+ nr = NULL,
+ na.rm = FALSE,
+ dec = 3,
+ perc = "",
+ filter = "top",
+ pageLength = 10,
+ dom = "",
+ style = "bootstrap4",
+ rownames = FALSE,
+ caption = NULL,
+ envir = parent.frame(),
+ ...
+)Arguments
+- object +
Data.frame to display
+
+
+- vars +
Variables to show (default is all)
+
+
+- filt +
Filter to apply to the specified dataset. For example "price > 10000" if dataset is "diamonds" (default is "")
+
+
+- arr +
Expression to arrange (sort) the data on (e.g., "color, desc(price)")
+
+
+- rows +
Select rows in the specified dataset. For example "1:10" for the first 10 rows or "n()-10:n()" for the last 10 rows (default is NULL)
+
+
+- nr +
Number of rows of data to include in the table. This function will be mainly used in reports so it is best to keep this number small
+
+
+- na.rm +
Remove rows with missing values (default is FALSE)
+
+
+- dec +
Number of decimal places to show. Default is no rounding (NULL)
+
+
+- perc +
Vector of column names to be displayed as a percentage
+
+
+- filter +
Show column filters in DT table. Options are "none", "top", "bottom"
+
+
+- pageLength +
Number of rows to show in table
+
+
+- dom +
Table control elements to show on the page. See https://datatables.net/reference/option/dom
+
+
+- style +
Table formatting style ("bootstrap" or "default")
+
+
+- rownames +
Show data.frame rownames. Default is FALSE
+
+
+- caption +
Table caption
+
+
+- envir +
Environment to extract data from
+
+
+- ... +
Additional arguments
+
+
Details
+View, search, sort, and filter a data.frame. For styling options see https://rstudio.github.io/DT/functions.html
+Examples
+if (FALSE) {
+dtab(mtcars)
+}
+
+Make an interactive table of summary statistics
+# S3 method for explore
+dtab(
+ object,
+ dec = 3,
+ searchCols = NULL,
+ order = NULL,
+ pageLength = NULL,
+ caption = NULL,
+ ...
+)Arguments
+- object +
Return value from
explore
+
+
+- dec +
Number of decimals to show
+
+
+- searchCols +
Column search and filter
+
+
+- order +
Column sorting
+
+
+- pageLength +
Page length
+
+
+- caption +
Table caption
+
+
+- ... +
further arguments passed to or from other methods
+
+
Details
+See https://radiant-rstats.github.io/docs/data/explore.html for an example in Radiant
+See also
+pivotr to create a pivot table
summary.pivotr to show summaries
Method to create datatables
+dtab(object, ...)Arguments
+- object +
Object of relevant class to render
+
+
+- ... +
Additional arguments
+
+
See also
+See dtab.data.frame to create an interactive table from a data.frame
See dtab.explore to create an interactive table from an explore object
See dtab.pivotr to create an interactive table from a pivotr object
Make an interactive pivot table
+# S3 method for pivotr
+dtab(
+ object,
+ format = "none",
+ perc = FALSE,
+ dec = 3,
+ searchCols = NULL,
+ order = NULL,
+ pageLength = NULL,
+ caption = NULL,
+ ...
+)Arguments
+- object +
Return value from
pivotr
+
+
+- format +
Show Color bar ("color_bar"), Heat map ("heat"), or None ("none")
+
+
+- perc +
Display numbers as percentages (TRUE or FALSE)
+
+
+- dec +
Number of decimals to show
+
+
+- searchCols +
Column search and filter
+
+
+- order +
Column sorting
+
+
+- pageLength +
Page length
+
+
+- caption +
Table caption
+
+
+- ... +
further arguments passed to or from other methods
+
+
Details
+See https://radiant-rstats.github.io/docs/data/pivotr.html for an example in Radiant
+See also
+pivotr to create the pivot table
summary.pivotr to print the table
Convert categorical variables to factors and deal with empty/missing values
+ Source:R/explore.R
+ empty_level.RdConvert categorical variables to factors and deal with empty/missing values
+empty_level(x)Arguments
+- x +
Categorical variable used in table
+
+
Value
+ + +Variable with updated levels
+Explore and summarize data
+explore(
+ dataset,
+ vars = "",
+ byvar = "",
+ fun = c("mean", "sd"),
+ top = "fun",
+ tabfilt = "",
+ tabsort = "",
+ tabslice = "",
+ nr = Inf,
+ data_filter = "",
+ arr = "",
+ rows = NULL,
+ envir = parent.frame()
+)Arguments
+- dataset +
Dataset to explore
+
+
+- vars +
(Numeric) variables to summarize
+
+
+- byvar +
Variable(s) to group data by
+
+
+- fun +
Functions to use for summarizing
+
+
+- top +
Use functions ("fun"), variables ("vars"), or group-by variables as column headers
+
+
+- tabfilt +
Expression used to filter the table (e.g., "Total > 10000")
+
+
+- tabsort +
Expression used to sort the table (e.g., "desc(Total)")
+
+
+- tabslice +
Expression used to filter table (e.g., "1:5")
+
+
+- nr +
Number of rows to display
+
+
+- data_filter +
Expression used to filter the dataset before creating the table (e.g., "price > 10000")
+
+
+- arr +
Expression to arrange (sort) the data on (e.g., "color, desc(price)")
+
+
+- rows +
Rows to select from the specified dataset
+
+
+- envir +
Environment to extract data from
+
+
Value
+ + +A list of all variables defined in the function as an object of class explore
+Details
+See https://radiant-rstats.github.io/docs/data/explore.html for an example in Radiant
+See also
+See summary.explore to show summaries
Examples
+explore(diamonds, c("price", "carat")) %>% str()
+#> List of 13
+#> $ tab :'data.frame': 2 obs. of 3 variables:
+#> ..$ variable: Factor w/ 2 levels "price","carat": 1 2
+#> ..$ mean : num [1:2] 3907.186 0.794
+#> ..$ sd : num [1:2] 3956.915 0.474
+#> ..- attr(*, "radiant_nrow")= int 2
+#> $ df_name : chr "diamonds"
+#> $ vars : chr [1:2] "price" "carat"
+#> $ byvar : NULL
+#> $ fun : chr [1:2] "mean" "sd"
+#> $ top : chr "fun"
+#> $ tabfilt : chr ""
+#> $ tabsort : chr ""
+#> $ tabslice : chr ""
+#> $ nr : num Inf
+#> $ data_filter: chr ""
+#> $ arr : chr ""
+#> $ rows : NULL
+#> - attr(*, "class")= chr [1:2] "explore" "list"
+explore(diamonds, "price:x")$tab
+#> variable mean sd
+#> 1 price 3.907186e+03 3956.9154001
+#> 2 carat 7.942833e-01 0.4738263
+#> 3 clarity 1.333333e-02 0.1147168
+#> 4 cut 3.366667e-02 0.1803998
+#> 5 color 1.273333e-01 0.3334016
+#> 6 depth 6.175267e+01 1.4460279
+#> 7 table 5.746533e+01 2.2411022
+#> 8 x 5.721823e+00 1.1240545
+explore(diamonds, c("price", "carat"), byvar = "cut", fun = c("n_missing", "skew"))$tab
+#> cut variable n_missing skew
+#> 1 Fair price 0 1.5741334
+#> 2 Fair carat 0 0.9285670
+#> 3 Good price 0 1.4885765
+#> 4 Good carat 0 1.0207909
+#> 5 Very Good price 0 1.6007752
+#> 6 Very Good carat 0 0.9370738
+#> 7 Premium price 0 1.4131786
+#> 8 Premium carat 0 0.9299567
+#> 9 Ideal price 0 1.7986601
+#> 10 Ideal carat 0 1.3654745
+
+Filter data with user-specified expression
+filter_data(dataset, filt = "", drop = TRUE)Arguments
+- dataset +
Data frame to filter
+
+
+- filt +
Filter expression to apply to the specified dataset
+
+
+- drop +
Drop unused factor levels after filtering (default is TRUE)
+
+
Value
+ + +Filtered data frame
+Details
+Filters can be used to view a sample from a selected dataset. For example, runif(nrow(.)) > .9 could be used to sample approximately 10
+Examples
+select(diamonds, 1:3) %>% filter_data(filt = "price > max(.$price) - 100")
+#> # A tibble: 2 × 3
+#> price carat clarity
+#> <int> <dbl> <fct>
+#> 1 18791 2.15 SI2
+#> 2 18745 2.36 SI2
+select(diamonds, 1:3) %>% filter_data(filt = "runif(nrow(.)) > .995")
+#> # A tibble: 13 × 3
+#> price carat clarity
+#> <int> <dbl> <fct>
+#> 1 1895 0.51 VVS2
+#> 2 2100 0.5 VS2
+#> 3 6062 1.06 SI1
+#> 4 1087 0.42 VS2
+#> 5 7091 1.12 VVS2
+#> 6 1240 0.4 VVS2
+#> 7 4155 1 SI2
+#> 8 4641 1 VS1
+#> 9 794 0.36 SI1
+#> 10 5939 1.01 SI1
+#> 11 4405 1.15 SI2
+#> 12 1300 0.38 IF
+#> 13 5483 1.13 SI1
+Find Dropbox folder
+find_dropbox(account = 1)Arguments
+- account +
Integer. If multiple accounts exist, specify which one to use. By default, the first account listed is used
+
+
Value
+ + +Path to Dropbox account
+Details
+Find the path for Dropbox if available
+Find Google Drive folder
+find_gdrive()Value
+ + +Path to Google Drive folder
+Details
+Find the path for Google Drive if available
+Find user directory
+find_home()Details
+Returns /Users/x and not /Users/x/Documents
+Find the Rstudio project folder
+find_project(mess = TRUE)Arguments
+- mess +
Show or hide messages (default mess = TRUE)
+
+
Value
+ + +Path to Rstudio project folder if available or else and empty string. The returned path is normalized
+Details
+Find the path for the Rstudio project folder if available. The returned path is normalized (see normalizePath)
Ensure column names are valid
+fix_names(x, lower = FALSE)Arguments
+- x +
Data.frame or vector of (column) names
+
+
+- lower +
Set letters to lower case (TRUE or FALSE)
+
+
Details
+Remove symbols, trailing and leading spaces, and convert to valid R column names. Opinionated version of make.names
Examples
+fix_names(c(" var-name ", "$amount spent", "100"))
+#> [1] "var_name" "amount_spent" "X100"
+Replace smart quotes etc.
+fix_smart(text, all = FALSE)Arguments
+- text +
Text to be parsed
+
+
+- all +
Should all non-ascii characters be removed? Default is FALSE
+
+
Flip the DT table to put Function, Variable, or Group by on top
+flip(expl, top = "fun")Arguments
+- expl +
Return value from
explore
+
+
+- top +
The variable (type) to display at the top of the table ("fun" for Function, "var" for Variable, and "byvar" for Group by. "fun" is the default
+
+
Details
+See https://radiant-rstats.github.io/docs/data/explore.html for an example in Radiant
+See also
+explore to calculate summaries
summary.explore to show summaries
dtab.explore to create the DT table
Examples
+explore(diamonds, "price:x", top = "var") %>% summary()
+#> Explore
+#> Data : diamonds
+#> Functions : mean, sd
+#> Top : Variables
+#>
+#> .function price carat clarity cut color depth table x
+#> mean 3,907.186 0.794 0.013 0.034 0.127 61.753 57.465 5.722
+#> sd 3,956.915 0.474 0.115 0.180 0.333 1.446 2.241 1.124
+explore(diamonds, "price", byvar = "cut", fun = c("n_obs", "skew"), top = "byvar") %>% summary()
+#> Explore
+#> Data : diamonds
+#> Grouped by : cut
+#> Functions : n_obs, skew
+#> Top : Group by
+#>
+#> variable function. Fair Good Very_Good Premium Ideal
+#> price n_obs 101.000 275.000 677.000 771.000 1,176.000
+#> price skew 1.574 1.489 1.601 1.413 1.799
+
+Format a data.frame with a specified number of decimal places
+ Source:R/radiant.R
+ format_df.RdFormat a data.frame with a specified number of decimal places
+format_df(tbl, dec = NULL, perc = FALSE, mark = "", na.rm = FALSE, ...)Arguments
+- tbl +
Data.frame
+
+
+- dec +
Number of decimals to show
+
+
+- perc +
Display numbers as percentages (TRUE or FALSE)
+
+
+- mark +
Thousand separator
+
+
+- na.rm +
Remove missing values
+
+
+- ... +
Additional arguments for format_nr
+
+
Value
+ + +Data.frame for printing
+Examples
+data.frame(x = c("a", "b"), y = c(1L, 2L), z = c(-0.0005, 3)) %>%
+ format_df(dec = 4)
+#> x y z
+#> 1 a 1 -0.0005
+#> 2 b 2 3.0000
+data.frame(x = c(1L, 2L), y = c(0.06, 0.8)) %>%
+ format_df(dec = 2, perc = TRUE)
+#> x y
+#> 1 1 6.00%
+#> 2 2 80.00%
+data.frame(x = c(1L, 2L, NA), y = c(NA, 1.008, 2.8)) %>%
+ format_df(dec = 2)
+#> x y
+#> 1 1 NA
+#> 2 2 1.01
+#> 3 NA 2.80
+Format a number with a specified number of decimal places, thousand sep, and a symbol
+ Source:R/radiant.R
+ format_nr.RdFormat a number with a specified number of decimal places, thousand sep, and a symbol
+format_nr(x, sym = "", dec = 2, perc = FALSE, mark = ",", na.rm = TRUE, ...)Arguments
+- x +
Number or vector
+
+
+- sym +
Symbol to use
+
+
+- dec +
Number of decimals to show
+
+
+- perc +
Display number as a percentage
+
+
+- mark +
Thousand separator
+
+
+- na.rm +
Remove missing values
+
+
+- ... +
Additional arguments passed to
formatC
+
+
Value
+ + +Character (vector) in the desired format
+Examples
+format_nr(2000, "$")
+#> [1] "$2,000.00"
+format_nr(2000, dec = 4)
+#> [1] "2,000.0000"
+format_nr(.05, perc = TRUE)
+#> [1] "5.00%"
+format_nr(c(.1, .99), perc = TRUE)
+#> [1] "10.00%" "99.00%"
+format_nr(data.frame(a = c(.1, .99)), perc = TRUE)
+#> [1] "10.00%" "99.00%"
+format_nr(data.frame(a = 1:10), sym = "$", dec = 0)
+#> [1] "$1" "$2" "$3" "$4" "$5" "$6" "$7" "$8" "$9" "$10"
+format_nr(c(1, 1.9, 1.008, 1.00))
+#> [1] "1.00" "1.90" "1.01" "1.00"
+format_nr(c(1, 1.9, 1.008, 1.00), drop0trailing = TRUE)
+#> [1] "1" "1.9" "1.01" "1"
+format_nr(NA)
+#> [1] ""
+format_nr(NULL)
+#> [1] ""
+Get variable class
+get_class(dat)Arguments
+- dat +
Dataset to evaluate
+
+
Value
+ + +Vector with class information for each variable
+Details
+Get variable class information for each column in a data.frame
+Examples
+get_class(mtcars)
+#> mpg cyl disp hp drat wt qsec vs
+#> "numeric" "numeric" "numeric" "numeric" "numeric" "numeric" "numeric" "numeric"
+#> am gear carb
+#> "numeric" "numeric" "numeric"
+Select variables and filter data
+get_data(
+ dataset,
+ vars = "",
+ filt = "",
+ arr = "",
+ rows = NULL,
+ data_view_rows = NULL,
+ na.rm = TRUE,
+ rev = FALSE,
+ envir = c()
+)Arguments
+- dataset +
Dataset or name of the data.frame
+
+
+- vars +
Variables to extract from the data.frame
+
+
+- filt +
Filter to apply to the specified dataset
+
+
+- arr +
Expression to use to arrange (sort) the specified dataset
+
+
+- rows +
Select rows in the specified dataset
+
+
+- data_view_rows +
Vector of rows to select. Only used by Data > View in Radiant. Users should use "rows" instead
+
+
+- na.rm +
Remove rows with missing values (default is TRUE)
+
+
+- rev +
Reverse filter and row selection (i.e., get the remainder)
+
+
+- envir +
Environment to extract data from
+
+
Value
+ + +Data.frame with specified columns and rows
+Details
+Function is used in radiant to select variables and filter data based on user input in string form
+Examples
+get_data(mtcars, vars = "cyl:vs", filt = "mpg > 25")
+#> cyl disp hp drat wt qsec vs
+#> Fiat 128 4 78.7 66 4.08 2.200 19.47 1
+#> Honda Civic 4 75.7 52 4.93 1.615 18.52 1
+#> Toyota Corolla 4 71.1 65 4.22 1.835 19.90 1
+#> Fiat X1-9 4 79.0 66 4.08 1.935 18.90 1
+#> Porsche 914-2 4 120.3 91 4.43 2.140 16.70 0
+#> Lotus Europa 4 95.1 113 3.77 1.513 16.90 1
+get_data(mtcars, vars = c("mpg", "cyl"), rows = 1:10)
+#> mpg cyl
+#> Mazda RX4 21.0 6
+#> Mazda RX4 Wag 21.0 6
+#> Datsun 710 22.8 4
+#> Hornet 4 Drive 21.4 6
+#> Hornet Sportabout 18.7 8
+#> Valiant 18.1 6
+#> Duster 360 14.3 8
+#> Merc 240D 24.4 4
+#> Merc 230 22.8 4
+#> Merc 280 19.2 6
+get_data(mtcars, vars = c("mpg", "cyl"), arr = "desc(mpg)", rows = "1:5")
+#> mpg cyl
+#> Toyota Corolla 33.9 4
+#> Fiat 128 32.4 4
+#> Honda Civic 30.4 4
+#> Lotus Europa 30.4 4
+#> Fiat X1-9 27.3 4
+Create data.frame summary
+get_summary(dataset, dc = get_class(dataset), dec = 3)Arguments
+- dataset +
Data.frame
+
+
+- dc +
Class for each variable
+
+
+- dec +
Number of decimals to show
+
+
Details
+Used in Radiant's Data > Transform tab
+Exporting glue_collapse from glue
+Details
+ +See glue::glue_collapse() in the glue package for more details
Exporting glue_data from glue
+Details
+ +See glue::glue_data() in the glue package for more details
Reference
+
+ Data > Manage+Functions used with Data > Manage + |
+ |
|---|---|
| + + | +Choose a directory interactively |
+
| + + | +Choose files interactively |
+
| + + | +Show dataset description |
+
| + + | +Find Dropbox folder |
+
| + + | +Find Google Drive folder |
+
| + + | +Find user directory |
+
| + + | +Find the Rstudio project folder |
+
| + + | +Ensure column names are valid |
+
| + + | +Select variables and filter data |
+
| + + | +Load data through clipboard on Windows or macOS |
+
| + + | +Parse file path into useful components |
+
| + + | +Generate code to read a file |
+
| + + | +Save data to clipboard on Windows or macOS |
+
| + + | +Workaround to store description file together with a parquet data file |
+
| + + | +Convert characters to factors |
+
+ Data > View+Functions used with Data > View + |
+ |
| + + | +Method to create datatables |
+
| + + | +Create an interactive table to view, search, sort, and filter data |
+
| + + | +Filter data with user-specified expression |
+
| + + | +Generate arrange commands from user input |
+
| + + | +Arrange data with user-specified expression |
+
| + + | +Slice data with user-specified expression |
+
| + + | +Search for a pattern in all columns of a data.frame |
+
| + + | +View data in a shiny-app |
+
+ Data > Visualize+Function used with Data > Visualize + |
+ |
| + + | +Visualize data using ggplot2 https://ggplot2.tidyverse.org/ |
+
| + + | +Create a qscatter plot similar to Stata |
+
| + + | +Work around to avoid (harmless) messages from ggplotly |
+
| + + | +Work around to avoid (harmless) messages from subplot |
+
+ Data > Pivot+Functions used with Data > Pivot + |
+ |
| + + | +Create a pivot table |
+
| + + | +Summary method for pivotr |
+
| + + | +Make an interactive pivot table |
+
| + + | +Plot method for the pivotr function |
+
+ Data > Explore+Functions used with Data > Pivot + |
+ |
| + + | +Explore and summarize data |
+
| + + | +Summary method for the explore function |
+
| + + | +Make an interactive table of summary statistics |
+
| + + | +Flip the DT table to put Function, Variable, or Group by on top |
+
+ Data > Transform+Functions used with Data > Transform + |
+ |
| + + | +Wrapper for as.character |
+
| + + | +Distance in kilometers or miles between two locations based on lat-long +Function based on http://www.movable-type.co.uk/scripts/latlong.html. Uses the haversine formula |
+
| + + | +Convert input in day-month-year format to date |
+
| + + | +Convert input in day-month-year-hour-minute format to date-time |
+
| + + | +Convert input in day-month-year-hour-minute-second format to date-time |
+
| + + | +Wrapper for lubridate's as.duration function. Result converted to numeric |
+
| + + | +Wrapper for factor with ordered = FALSE |
+
| + + | +Convert input in hour-minute format to time |
+
| + + | +Convert input in hour-minute-second format to time |
+
| + + | +Convert variable to integer avoiding potential issues with factors |
+
| + + | +Convert input in month-day-year format to date |
+
| + + | +Convert input in month-day-year-hour-minute format to date-time |
+
| + + | +Convert input in month-day-year-hour-minute-second format to date-time |
+
| + + | +Convert variable to numeric avoiding potential issues with factors |
+
| + + | +Convert input in year-month-day format to date |
+
| + + | +Convert input in year-month-day-hour-minute format to date-time |
+
| + + | +Convert input in year-month-day-hour-minute-second format to date-time |
+
| + + | +Center |
+
| + + | +Coefficient of variation |
+
| + + | +Calculate inverse of a variable |
+
| + + | +Is a variable empty |
+
| + + | +Convenience function for is.null or is.na |
+
| + + | +Is input a double (and not a date type)? |
+
| + + | +Is input a string? |
+
| + + | +Generate list of levels and unique values |
+
| + + | +Natural log |
+
| + + | +Generate a variable used to selected a training sample |
+
| + + | +Add ordered argument to lubridate::month |
+
| + + | +Add transformed variables to a data frame with the option to include a custom variable name extension |
+
| + + | +Number of missing values |
+
| + + | +Number of observations |
+
| + + | +Normalize a variable x by a variable y |
+
| + + | +Convert a string of numbers into a vector |
+
| + + | +Margin of error |
+
| + + | +Margin of error for proportion |
+
| + + | +Calculate the mode (modal value) and return a label |
+
|
+
|
+ Calculate percentiles |
+
| + + | +Calculate proportion |
+
| + + | +Remove/reorder levels |
+
| + + | +Standard deviation for the population |
+
| + + | +Standard deviation for proportion |
+
| + + | +Standard error |
+
| + + | +Standard error for proportion |
+
| + + | +Show all rows with duplicated values (not just the first or last) |
+
| + + | +Calculate square of a variable |
+
| + + | +Standardize |
+
| + + | +Method to store variables in a dataset in Radiant |
+
| + + | +Create data.frame from a table |
+
| + + | +Variance for the population |
+
| + + | +Variance for proportion |
+
| + + | +Add ordered argument to lubridate::wday |
+
| + + | +Weighted standard deviation |
+
| + + | +Index of the maximum per row |
+
| + + | +Index of the minimum per row |
+
|
+
|
+ Summarize a set of numeric vectors per row |
+
| + + | +Split a numeric variable into a number of bins and return a vector of bin numbers |
+
+ Data > Combine+Functions used with Data > Combine + |
+ |
| + + | +Combine datasets using dplyr's bind and join functions |
+
+ Report+Functions used with Report > Rmd and Report > R + |
+ |
| + + | +Replace smart quotes etc. |
+
| + + | +Format a data.frame with a specified number of decimal places |
+
| + + | +Format a number with a specified number of decimal places, thousand sep, and a symbol |
+
| + + | +Round doubles in a data.frame to a specified number of decimal places |
+
| + + | +Register a data.frame or list in Radiant |
+
| + + | +Deregister a data.frame or list in Radiant |
+
| + + | +Base method used to render htmlwidgets |
+
| + + | +Method to render DT tables |
+
| + + | +Method to render plotly plots |
+
+ Convenience functions+Convenience functions + |
+ |
| + + | +Convenience function to add a class |
+
| + + | +Convenience function to add a markdown description to a data.frame |
+
| + + | +Get variable class |
+
| + + | +Labels for confidence intervals |
+
| + + | +Values at confidence levels |
+
| + + | +Source all package functions |
+
| + + | +Copy attributes from one object to another |
+
| + + | +Source for package functions |
+
| + + | +Does a vector have non-zero variability? |
+
| + + | +Convert categorical variables to factors and deal with empty/missing values |
+
| + + | +Create data.frame summary |
+
| + + | +Find index corrected for missing values and filters |
+
| + + | +Install webshot and phantomjs |
+
| + + | +Create a vector of interaction terms for linear and logistic regression |
+
| + + | +Create a vector of quadratic and cubed terms for use in linear and logistic regression |
+
| + + | +Alias used to add an attribute |
+
| + + | +Add stars based on p.values |
+
| + + | +Hide warnings and messages and return invisible |
+
| + + | +Hide warnings and messages and return result |
+
+ Starting radiant.data+Functions used to start radiant shiny apps + |
+ |
| + + | +Launch radiant apps |
+
| + + | +radiant.data |
+
| + + | +Start radiant.data app but do not open a browser |
+
| + + | +Launch the radiant.data app in the Rstudio viewer |
+
| + + | +Launch the radiant.data app in an Rstudio window |
+
+ Re-exported+Functions exported from other packages + |
+ |
|
+
|
+ Objects exported from other packages |
+
+ Data sets+Data sets bundled with radiant.data + |
+ |
| + + | +Avengers |
+
| + + | +Diamond prices |
+
| + + | +Comic publishers |
+
| + + | +Super heroes |
+
| + + | +Survival data for the Titanic |
+
+ Deprecated+Deprecated + |
+ |
| + + | +Deprecated function(s) in the radiant.data package |
+
| + + | +Deprecated: Store method for the pivotr function |
+
| + + | +Deprecated: Store method for the explore function |
+
Find index corrected for missing values and filters
+indexr(dataset, vars = "", filt = "", arr = "", rows = NULL, cmd = "")Arguments
+- dataset +
Dataset
+
+
+- vars +
Variables to select
+
+
+- filt +
Data filter
+
+
+- arr +
Expression to arrange (sort) the data on (e.g., "color, desc(price)")
+
+
+- rows +
Selected rows
+
+
+- cmd +
A command used to customize the data
+
+
Install webshot and phantomjs
+install_webshot()Calculate inverse of a variable
+inverse(x)Arguments
+- x +
Input variable
+
+
Value
+ + +1/x
+Is a variable empty
+is.empty(x, empty = "\\s*")Arguments
+- x +
Character value to evaluate
+
+
+- empty +
Indicate what 'empty' means. Default is empty string (i.e., "")
+
+
Value
+ + +TRUE if empty, else FALSE
+Details
+Is a variable empty
+Examples
+is.empty("")
+#> [1] TRUE
+is.empty(NULL)
+#> [1] TRUE
+is.empty(NA)
+#> [1] TRUE
+is.empty(c())
+#> [1] TRUE
+is.empty("none", empty = "none")
+#> [1] TRUE
+is.empty("")
+#> [1] TRUE
+is.empty(" ")
+#> [1] TRUE
+is.empty(" something ")
+#> [1] FALSE
+is.empty(c("", "something"))
+#> [1] FALSE
+is.empty(c(NA, 1:100))
+#> [1] FALSE
+is.empty(mtcars)
+#> [1] FALSE
+Is input a double (and not a date type)?
+is_double(x)Arguments
+- x +
Input
+
+
Value
+ + +TRUE if double and not a type of date, else FALSE
+Is a character variable defined
+is_empty(x, empty = "\\s*")Arguments
+- x +
Character value to evaluate
+- empty +
Indicate what 'empty' means. Default is empty string (i.e., "")
+
Value
+TRUE if empty, else FALSE
+Details
+Is a variable NULL or an empty string
+Examples
+is_empty("")
+#> [1] TRUE
+is_empty(NULL)
+#> [1] TRUE
+is_empty(NA)
+#> [1] TRUE
+is_empty(c())
+#> [1] TRUE
+is_empty("none", empty = "none")
+#> [1] TRUE
+is_empty("")
+#> [1] TRUE
+is_empty(" ")
+#> [1] TRUE
+is_empty(" something ")
+#> [1] FALSE
+is_empty(c("", "something"))
+#> [1] FALSE
+is_empty(c(NA, 1:100))
+#> [1] FALSE
+is_empty(mtcars)
+#> [1] FALSE
+Convenience function for is.null or is.na
+is_not(x)Arguments
+- x +
Input
+
+
Examples
+is_not(NA)
+#> [1] TRUE
+is_not(NULL)
+#> [1] TRUE
+is_not(c())
+#> [1] TRUE
+is_not(list())
+#> [1] TRUE
+is_not(data.frame())
+#> [1] TRUE
+Is input a string?
+is_string(x)Arguments
+- x +
Input
+
+
Value
+ + +TRUE if string, else FALSE
+Examples
+is_string(" ")
+#> [1] FALSE
+is_string("data")
+#> [1] TRUE
+is_string(c("data", ""))
+#> [1] FALSE
+is_string(NULL)
+#> [1] FALSE
+is_string(NA)
+#> [1] FALSE
+Create a vector of interaction terms for linear and logistic regression
+ Source:R/radiant.R
+ iterms.RdCreate a vector of interaction terms for linear and logistic regression
+iterms(vars, nway = 2, sep = ":")Arguments
+- vars +
Labels to use
+
+
+- nway +
2-way (2) or 3-way (3) interaction labels to create
+
+
+- sep +
Separator to use between variable names (e.g., :)
+
+
Value
+ + +Character vector of interaction term labels
+Exporting knit_print from knitr
+Details
+ +See knit_print in the knitr package for more details
Launch radiant apps
+launch(package = "radiant.data", run = "viewer", state, ...)Arguments
+- package +
Radiant package to start. One of "radiant.data", "radiant.design", "radiant.basics", "radiant.model", "radiant.multivariate", or "radiant"
+
+
+- run +
Run a radiant app in an external browser ("browser"), an Rstudio window ("window"), or in the Rstudio viewer ("viewer")
+
+
+- state +
Path to statefile to load
+
+
+- ... +
additional arguments to pass to shiny::runApp (e.g, port = 8080)
+
+
Details
+See https://radiant-rstats.github.io/docs/ for radiant documentation and tutorials
+Examples
+if (FALSE) {
+launch()
+launch(run = "viewer")
+launch(run = "window")
+launch(run = "browser")
+}
+
+Generate list of levels and unique values
+level_list(dataset, ...)Arguments
+- dataset +
A data.frame
+
+
+- ... +
Unquoted variable names to evaluate
+
+
Examples
+data.frame(a = c(rep("a", 5), rep("b", 5)), b = c(rep(1, 5), 6:10)) %>% level_list()
+#> $a
+#> [1] "a" "b"
+#>
+#> $b
+#> [1] 1 6 7 8 9 10
+#>
+level_list(mtcars, mpg, cyl)
+#> $mpg
+#> [1] 21.0 22.8 21.4 18.7 18.1 14.3 24.4 19.2 17.8 16.4 17.3 15.2 10.4 14.7 32.4
+#> [16] 30.4 33.9 21.5 15.5 13.3 27.3 26.0 15.8 19.7 15.0
+#>
+#> $cyl
+#> [1] 6 4 8
+#>
+
+Natural log
+ln(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
Remove missing values (default is TRUE)
+
+
Value
+ + +Natural log of vector
+Examples
+ln(runif(10, 1, 2))
+#> [1] 0.4846652 0.6214164 0.5995395 0.4299635 0.3771382 0.2102091 0.5922178
+#> [8] 0.2149426 0.5359432 0.2461529
+
+Load data through clipboard on Windows or macOS
+load_clip(delim = "\t", text, suppress = TRUE)Arguments
+- delim +
Delimiter to use (tab is the default)
+
+
+- text +
Text input to convert to table
+
+
+- suppress +
Suppress warnings
+
+
Details
+Extract data from the clipboard into a data.frame on Windows or macOS
+See also
+See the save_clip
Generate arrange commands from user input
+make_arrange_cmd(expr, dataset = "")Arguments
+- expr +
Expression to use arrange rows from the specified dataset
+
+
+- dataset +
String with dataset name
+
+
Value
+ + +Arrange command
+Details
+Form arrange command from user input
+Generate a variable used to selected a training sample
+make_train(n = 0.7, nr = NULL, blocks = NULL, seed = 1234)Arguments
+- n +
Number (or fraction) of observations to label as training
+
+
+- nr +
Number of rows in the dataset
+
+
+- blocks +
A vector to use for blocking or a data.frame from which to construct a blocking vector
+
+
+- seed +
Random seed
+
+
Value
+ + +0/1 variables for filtering
+Examples
+make_train(.5, 10)
+#> [1] 1 1 0 0 0 1 0 1 1 0
+make_train(.5, 10) %>% table()
+#> .
+#> 0 1
+#> 5 5
+make_train(100, 1000) %>% table()
+#> .
+#> 0 1
+#> 900 100
+make_train(.15, blocks = mtcars$vs) %>% table() / nrow(mtcars)
+#> .
+#> 0 1
+#> 0.84375 0.15625
+make_train(.10, blocks = iris$Species) %>% table() / nrow(iris)
+#> .
+#> 0 1
+#> 0.9 0.1
+make_train(.5, blocks = iris[, c("Petal.Width", "Species")]) %>% table()
+#> .
+#> 0 1
+#> 75 75
+
+Convert a string of numbers into a vector
+make_vec(x)Arguments
+- x +
A string of numbers that may include fractions
+
+
Examples
+make_vec("1 2 4")
+#> [1] 1 2 4
+make_vec("1/2 2/3 4/5")
+#> [1] 1/2 2/3 4/5
+make_vec(0.1)
+#> [1] 0.1
+Margin of error
+me(x, conf_lev = 0.95, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- conf_lev +
Confidence level. The default is 0.95
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Margin of error
+Examples
+me(rnorm(100))
+#> [1] 0.2048265
+
+Margin of error for proportion
+meprop(x, conf_lev = 0.95, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- conf_lev +
Confidence level. The default is 0.95
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Margin of error
+Calculate the mode (modal value) and return a label
+modal(x, na.rm = TRUE)Arguments
+- x +
A vector
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Details
+From https://www.tutorialspoint.com/r/r_mean_median_mode.htm
+Add ordered argument to lubridate::month
+month(x, label = FALSE, abbr = TRUE, ordered = FALSE)Arguments
+- x +
Input date vector
+
+
+- label +
Month as label (TRUE, FALSE)
+
+
+- abbr +
Abbreviate label (TRUE, FALSE)
+
+
+- ordered +
Order factor (TRUE, FALSE)
+
+
See also
+See the month function in the lubridate package for additional details
Add transformed variables to a data frame with the option to include a custom variable name extension
+ Source:R/transform.R
+ mutate_ext.RdAdd transformed variables to a data frame with the option to include a custom variable name extension
+mutate_ext(.tbl, .funs, ..., .ext = "", .vars = c())Arguments
+- .tbl +
Data frame to add transformed variables to
+
+
+- .funs +
Function(s) to apply (e.g., log)
+
+
+- ... +
Variables to transform
+
+
+- .ext +
Extension to add for each variable
+
+
+- .vars +
A list of columns generated by dplyr::vars(), or a character vector of column names, or a numeric vector of column positions.
+
+
Details
+Wrapper for dplyr::mutate_at that allows custom variable name extensions
+Examples
+mutate_ext(mtcars, .funs = log, mpg, cyl, .ext = "_ln")
+#> mpg cyl disp hp drat wt qsec vs am gear carb
+#> Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
+#> Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
+#> Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
+#> Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
+#> Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
+#> Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
+#> Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4
+#> Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
+#> Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
+#> Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
+#> Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
+#> Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3
+#> Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
+#> Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3
+#> Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4
+#> Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4
+#> Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
+#> Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
+#> Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
+#> Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
+#> Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
+#> Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2
+#> AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
+#> Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4
+#> Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2
+#> Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
+#> Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
+#> Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
+#> Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4
+#> Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6
+#> Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8
+#> Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2
+#> mpg_ln cyl_ln
+#> Mazda RX4 3.044522 1.791759
+#> Mazda RX4 Wag 3.044522 1.791759
+#> Datsun 710 3.126761 1.386294
+#> Hornet 4 Drive 3.063391 1.791759
+#> Hornet Sportabout 2.928524 2.079442
+#> Valiant 2.895912 1.791759
+#> Duster 360 2.660260 2.079442
+#> Merc 240D 3.194583 1.386294
+#> Merc 230 3.126761 1.386294
+#> Merc 280 2.954910 1.791759
+#> Merc 280C 2.879198 1.791759
+#> Merc 450SE 2.797281 2.079442
+#> Merc 450SL 2.850707 2.079442
+#> Merc 450SLC 2.721295 2.079442
+#> Cadillac Fleetwood 2.341806 2.079442
+#> Lincoln Continental 2.341806 2.079442
+#> Chrysler Imperial 2.687847 2.079442
+#> Fiat 128 3.478158 1.386294
+#> Honda Civic 3.414443 1.386294
+#> Toyota Corolla 3.523415 1.386294
+#> Toyota Corona 3.068053 1.386294
+#> Dodge Challenger 2.740840 2.079442
+#> AMC Javelin 2.721295 2.079442
+#> Camaro Z28 2.587764 2.079442
+#> Pontiac Firebird 2.954910 2.079442
+#> Fiat X1-9 3.306887 1.386294
+#> Porsche 914-2 3.258097 1.386294
+#> Lotus Europa 3.414443 1.386294
+#> Ford Pantera L 2.760010 2.079442
+#> Ferrari Dino 2.980619 1.791759
+#> Maserati Bora 2.708050 2.079442
+#> Volvo 142E 3.063391 1.386294
+mutate_ext(mtcars, .funs = log, .ext = "_ln")
+#> mpg cyl disp hp drat wt qsec vs am gear carb
+#> Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
+#> Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
+#> Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
+#> Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
+#> Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
+#> Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
+#> Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4
+#> Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
+#> Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
+#> Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
+#> Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
+#> Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3
+#> Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
+#> Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3
+#> Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4
+#> Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4
+#> Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
+#> Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
+#> Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
+#> Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
+#> Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
+#> Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2
+#> AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
+#> Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4
+#> Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2
+#> Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
+#> Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
+#> Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
+#> Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4
+#> Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6
+#> Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8
+#> Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2
+#> mpg_ln cyl_ln disp_ln hp_ln drat_ln wt_ln
+#> Mazda RX4 3.044522 1.791759 5.075174 4.700480 1.360977 0.9631743
+#> Mazda RX4 Wag 3.044522 1.791759 5.075174 4.700480 1.360977 1.0560527
+#> Datsun 710 3.126761 1.386294 4.682131 4.532599 1.348073 0.8415672
+#> Hornet 4 Drive 3.063391 1.791759 5.552960 4.700480 1.124930 1.1678274
+#> Hornet Sportabout 2.928524 2.079442 5.886104 5.164786 1.147402 1.2354715
+#> Valiant 2.895912 1.791759 5.416100 4.653960 1.015231 1.2412686
+#> Duster 360 2.660260 2.079442 5.886104 5.501258 1.166271 1.2725656
+#> Merc 240D 3.194583 1.386294 4.988390 4.127134 1.305626 1.1600209
+#> Merc 230 3.126761 1.386294 4.947340 4.553877 1.366092 1.1474025
+#> Merc 280 2.954910 1.791759 5.121580 4.812184 1.366092 1.2354715
+#> Merc 280C 2.879198 1.791759 5.121580 4.812184 1.366092 1.2354715
+#> Merc 450SE 2.797281 2.079442 5.619676 5.192957 1.121678 1.4036430
+#> Merc 450SL 2.850707 2.079442 5.619676 5.192957 1.121678 1.3164082
+#> Merc 450SLC 2.721295 2.079442 5.619676 5.192957 1.121678 1.3297240
+#> Cadillac Fleetwood 2.341806 2.079442 6.156979 5.323010 1.075002 1.6582281
+#> Lincoln Continental 2.341806 2.079442 6.131226 5.370638 1.098612 1.6908336
+#> Chrysler Imperial 2.687847 2.079442 6.086775 5.438079 1.172482 1.6761615
+#> Fiat 128 3.478158 1.386294 4.365643 4.189655 1.406097 0.7884574
+#> Honda Civic 3.414443 1.386294 4.326778 3.951244 1.595339 0.4793350
+#> Toyota Corolla 3.523415 1.386294 4.264087 4.174387 1.439835 0.6070445
+#> Toyota Corona 3.068053 1.386294 4.788325 4.574711 1.308333 0.9021918
+#> Dodge Challenger 2.740840 2.079442 5.762051 5.010635 1.015231 1.2584610
+#> AMC Javelin 2.721295 2.079442 5.717028 5.010635 1.147402 1.2340169
+#> Camaro Z28 2.587764 2.079442 5.857933 5.501258 1.316408 1.3454724
+#> Pontiac Firebird 2.954910 2.079442 5.991465 5.164786 1.124930 1.3467736
+#> Fiat X1-9 3.306887 1.386294 4.369448 4.189655 1.406097 0.6601073
+#> Porsche 914-2 3.258097 1.386294 4.789989 4.510860 1.488400 0.7608058
+#> Lotus Europa 3.414443 1.386294 4.554929 4.727388 1.327075 0.4140944
+#> Ford Pantera L 2.760010 2.079442 5.860786 5.575949 1.439835 1.1537316
+#> Ferrari Dino 2.980619 1.791759 4.976734 5.164786 1.286474 1.0188473
+#> Maserati Bora 2.708050 2.079442 5.707110 5.814131 1.264127 1.2725656
+#> Volvo 142E 3.063391 1.386294 4.795791 4.691348 1.413423 1.0224509
+#> qsec_ln vs_ln am_ln gear_ln carb_ln
+#> Mazda RX4 2.800933 -Inf 0 1.386294 1.3862944
+#> Mazda RX4 Wag 2.834389 -Inf 0 1.386294 1.3862944
+#> Datsun 710 2.923699 0 0 1.386294 0.0000000
+#> Hornet 4 Drive 2.967333 0 -Inf 1.098612 0.0000000
+#> Hornet Sportabout 2.834389 -Inf -Inf 1.098612 0.6931472
+#> Valiant 3.006672 0 -Inf 1.098612 0.0000000
+#> Duster 360 2.762538 -Inf -Inf 1.098612 1.3862944
+#> Merc 240D 2.995732 0 -Inf 1.386294 0.6931472
+#> Merc 230 3.131137 0 -Inf 1.386294 0.6931472
+#> Merc 280 2.906901 0 -Inf 1.386294 1.3862944
+#> Merc 280C 2.939162 0 -Inf 1.386294 1.3862944
+#> Merc 450SE 2.856470 -Inf -Inf 1.098612 1.0986123
+#> Merc 450SL 2.867899 -Inf -Inf 1.098612 1.0986123
+#> Merc 450SLC 2.890372 -Inf -Inf 1.098612 1.0986123
+#> Cadillac Fleetwood 2.889260 -Inf -Inf 1.098612 1.3862944
+#> Lincoln Continental 2.880321 -Inf -Inf 1.098612 1.3862944
+#> Chrysler Imperial 2.857619 -Inf -Inf 1.098612 1.3862944
+#> Fiat 128 2.968875 0 0 1.386294 0.0000000
+#> Honda Civic 2.918851 0 0 1.386294 0.6931472
+#> Toyota Corolla 2.990720 0 0 1.386294 0.0000000
+#> Toyota Corona 2.996232 0 -Inf 1.098612 0.0000000
+#> Dodge Challenger 2.825537 -Inf -Inf 1.098612 0.6931472
+#> AMC Javelin 2.850707 -Inf -Inf 1.098612 0.6931472
+#> Camaro Z28 2.735017 -Inf -Inf 1.098612 1.3862944
+#> Pontiac Firebird 2.836150 -Inf -Inf 1.098612 0.6931472
+#> Fiat X1-9 2.939162 0 0 1.386294 0.0000000
+#> Porsche 914-2 2.815409 -Inf 0 1.609438 0.6931472
+#> Lotus Europa 2.827314 0 0 1.609438 0.6931472
+#> Ford Pantera L 2.674149 -Inf 0 1.609438 1.3862944
+#> Ferrari Dino 2.740840 -Inf 0 1.609438 1.7917595
+#> Maserati Bora 2.681022 -Inf 0 1.609438 2.0794415
+#> Volvo 142E 2.923162 0 0 1.386294 0.6931472
+mutate_ext(mtcars, .funs = log)
+#> mpg cyl disp hp drat wt
+#> Mazda RX4 3.044522 1.791759 5.075174 4.700480 1.360977 0.9631743
+#> Mazda RX4 Wag 3.044522 1.791759 5.075174 4.700480 1.360977 1.0560527
+#> Datsun 710 3.126761 1.386294 4.682131 4.532599 1.348073 0.8415672
+#> Hornet 4 Drive 3.063391 1.791759 5.552960 4.700480 1.124930 1.1678274
+#> Hornet Sportabout 2.928524 2.079442 5.886104 5.164786 1.147402 1.2354715
+#> Valiant 2.895912 1.791759 5.416100 4.653960 1.015231 1.2412686
+#> Duster 360 2.660260 2.079442 5.886104 5.501258 1.166271 1.2725656
+#> Merc 240D 3.194583 1.386294 4.988390 4.127134 1.305626 1.1600209
+#> Merc 230 3.126761 1.386294 4.947340 4.553877 1.366092 1.1474025
+#> Merc 280 2.954910 1.791759 5.121580 4.812184 1.366092 1.2354715
+#> Merc 280C 2.879198 1.791759 5.121580 4.812184 1.366092 1.2354715
+#> Merc 450SE 2.797281 2.079442 5.619676 5.192957 1.121678 1.4036430
+#> Merc 450SL 2.850707 2.079442 5.619676 5.192957 1.121678 1.3164082
+#> Merc 450SLC 2.721295 2.079442 5.619676 5.192957 1.121678 1.3297240
+#> Cadillac Fleetwood 2.341806 2.079442 6.156979 5.323010 1.075002 1.6582281
+#> Lincoln Continental 2.341806 2.079442 6.131226 5.370638 1.098612 1.6908336
+#> Chrysler Imperial 2.687847 2.079442 6.086775 5.438079 1.172482 1.6761615
+#> Fiat 128 3.478158 1.386294 4.365643 4.189655 1.406097 0.7884574
+#> Honda Civic 3.414443 1.386294 4.326778 3.951244 1.595339 0.4793350
+#> Toyota Corolla 3.523415 1.386294 4.264087 4.174387 1.439835 0.6070445
+#> Toyota Corona 3.068053 1.386294 4.788325 4.574711 1.308333 0.9021918
+#> Dodge Challenger 2.740840 2.079442 5.762051 5.010635 1.015231 1.2584610
+#> AMC Javelin 2.721295 2.079442 5.717028 5.010635 1.147402 1.2340169
+#> Camaro Z28 2.587764 2.079442 5.857933 5.501258 1.316408 1.3454724
+#> Pontiac Firebird 2.954910 2.079442 5.991465 5.164786 1.124930 1.3467736
+#> Fiat X1-9 3.306887 1.386294 4.369448 4.189655 1.406097 0.6601073
+#> Porsche 914-2 3.258097 1.386294 4.789989 4.510860 1.488400 0.7608058
+#> Lotus Europa 3.414443 1.386294 4.554929 4.727388 1.327075 0.4140944
+#> Ford Pantera L 2.760010 2.079442 5.860786 5.575949 1.439835 1.1537316
+#> Ferrari Dino 2.980619 1.791759 4.976734 5.164786 1.286474 1.0188473
+#> Maserati Bora 2.708050 2.079442 5.707110 5.814131 1.264127 1.2725656
+#> Volvo 142E 3.063391 1.386294 4.795791 4.691348 1.413423 1.0224509
+#> qsec vs am gear carb
+#> Mazda RX4 2.800933 -Inf 0 1.386294 1.3862944
+#> Mazda RX4 Wag 2.834389 -Inf 0 1.386294 1.3862944
+#> Datsun 710 2.923699 0 0 1.386294 0.0000000
+#> Hornet 4 Drive 2.967333 0 -Inf 1.098612 0.0000000
+#> Hornet Sportabout 2.834389 -Inf -Inf 1.098612 0.6931472
+#> Valiant 3.006672 0 -Inf 1.098612 0.0000000
+#> Duster 360 2.762538 -Inf -Inf 1.098612 1.3862944
+#> Merc 240D 2.995732 0 -Inf 1.386294 0.6931472
+#> Merc 230 3.131137 0 -Inf 1.386294 0.6931472
+#> Merc 280 2.906901 0 -Inf 1.386294 1.3862944
+#> Merc 280C 2.939162 0 -Inf 1.386294 1.3862944
+#> Merc 450SE 2.856470 -Inf -Inf 1.098612 1.0986123
+#> Merc 450SL 2.867899 -Inf -Inf 1.098612 1.0986123
+#> Merc 450SLC 2.890372 -Inf -Inf 1.098612 1.0986123
+#> Cadillac Fleetwood 2.889260 -Inf -Inf 1.098612 1.3862944
+#> Lincoln Continental 2.880321 -Inf -Inf 1.098612 1.3862944
+#> Chrysler Imperial 2.857619 -Inf -Inf 1.098612 1.3862944
+#> Fiat 128 2.968875 0 0 1.386294 0.0000000
+#> Honda Civic 2.918851 0 0 1.386294 0.6931472
+#> Toyota Corolla 2.990720 0 0 1.386294 0.0000000
+#> Toyota Corona 2.996232 0 -Inf 1.098612 0.0000000
+#> Dodge Challenger 2.825537 -Inf -Inf 1.098612 0.6931472
+#> AMC Javelin 2.850707 -Inf -Inf 1.098612 0.6931472
+#> Camaro Z28 2.735017 -Inf -Inf 1.098612 1.3862944
+#> Pontiac Firebird 2.836150 -Inf -Inf 1.098612 0.6931472
+#> Fiat X1-9 2.939162 0 0 1.386294 0.0000000
+#> Porsche 914-2 2.815409 -Inf 0 1.609438 0.6931472
+#> Lotus Europa 2.827314 0 0 1.609438 0.6931472
+#> Ford Pantera L 2.674149 -Inf 0 1.609438 1.3862944
+#> Ferrari Dino 2.740840 -Inf 0 1.609438 1.7917595
+#> Maserati Bora 2.681022 -Inf 0 1.609438 2.0794415
+#> Volvo 142E 2.923162 0 0 1.386294 0.6931472
+mutate_ext(mtcars, .funs = log, .ext = "_ln", .vars = vars(mpg, cyl))
+#> mpg cyl disp hp drat wt qsec vs am gear carb
+#> Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
+#> Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
+#> Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
+#> Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
+#> Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
+#> Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
+#> Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4
+#> Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
+#> Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
+#> Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
+#> Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
+#> Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3
+#> Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
+#> Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3
+#> Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4
+#> Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4
+#> Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
+#> Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
+#> Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
+#> Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
+#> Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
+#> Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2
+#> AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
+#> Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4
+#> Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2
+#> Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
+#> Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
+#> Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
+#> Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4
+#> Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6
+#> Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8
+#> Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2
+#> mpg_ln cyl_ln
+#> Mazda RX4 3.044522 1.791759
+#> Mazda RX4 Wag 3.044522 1.791759
+#> Datsun 710 3.126761 1.386294
+#> Hornet 4 Drive 3.063391 1.791759
+#> Hornet Sportabout 2.928524 2.079442
+#> Valiant 2.895912 1.791759
+#> Duster 360 2.660260 2.079442
+#> Merc 240D 3.194583 1.386294
+#> Merc 230 3.126761 1.386294
+#> Merc 280 2.954910 1.791759
+#> Merc 280C 2.879198 1.791759
+#> Merc 450SE 2.797281 2.079442
+#> Merc 450SL 2.850707 2.079442
+#> Merc 450SLC 2.721295 2.079442
+#> Cadillac Fleetwood 2.341806 2.079442
+#> Lincoln Continental 2.341806 2.079442
+#> Chrysler Imperial 2.687847 2.079442
+#> Fiat 128 3.478158 1.386294
+#> Honda Civic 3.414443 1.386294
+#> Toyota Corolla 3.523415 1.386294
+#> Toyota Corona 3.068053 1.386294
+#> Dodge Challenger 2.740840 2.079442
+#> AMC Javelin 2.721295 2.079442
+#> Camaro Z28 2.587764 2.079442
+#> Pontiac Firebird 2.954910 2.079442
+#> Fiat X1-9 3.306887 1.386294
+#> Porsche 914-2 3.258097 1.386294
+#> Lotus Europa 3.414443 1.386294
+#> Ford Pantera L 2.760010 2.079442
+#> Ferrari Dino 2.980619 1.791759
+#> Maserati Bora 2.708050 2.079442
+#> Volvo 142E 3.063391 1.386294
+
+Number of missing values
+n_missing(x, ...)Arguments
+- x +
Input variable
+
+
+- ... +
Additional arguments
+
+
Value
+ + +number of missing values
+Examples
+n_missing(c("a", "b", NA))
+#> [1] 1
+
+Number of observations
+n_obs(x, ...)Arguments
+- x +
Input variable
+
+
+- ... +
Additional arguments
+
+
Value
+ + +number of observations
+Examples
+n_obs(c("a", "b", NA))
+#> [1] 3
+
+Normalize a variable x by a variable y
+normalize(x, y)Arguments
+- x +
Input variable
+
+
+- y +
Normalizing variable
+
+
Value
+ + +x/y
+Parse file path into useful components
+parse_path(path, chr = "", pdir = getwd(), mess = TRUE)Arguments
+- path +
Path to be parsed
+
+
+- chr +
Character to wrap around path for display
+
+
+- pdir +
Project directory if available
+
+
+- mess +
Print messages if Dropbox or Google Drive not found
+
+
Details
+Parse file path into useful components (i.e., file name, file extension, relative path, etc.)
+Examples
+list.files(".", full.names = TRUE)[1] %>% parse_path()
+#> $path
+#> [1] "/Users/vnijs/gh/radiant.data/docs/reference/Rplot001.png"
+#>
+#> $rpath
+#> Rplot001.png
+#>
+#> $filename
+#> [1] "Rplot001.png"
+#>
+#> $fext
+#> [1] "png"
+#>
+#> $objname
+#> [1] "Rplot001"
+#>
+Calculate percentiles
+p01(x, na.rm = TRUE)
+
+p025(x, na.rm = TRUE)
+
+p05(x, na.rm = TRUE)
+
+p10(x, na.rm = TRUE)
+
+p25(x, na.rm = TRUE)
+
+p75(x, na.rm = TRUE)
+
+p90(x, na.rm = TRUE)
+
+p95(x, na.rm = TRUE)
+
+p975(x, na.rm = TRUE)
+
+p99(x, na.rm = TRUE)Arguments
+- x +
Numeric vector
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Examples
+p01(0:100)
+#> 1%
+#> 1
+
+Summarize a set of numeric vectors per row
+pfun(..., fun, na.rm = TRUE)
+
+psum(..., na.rm = TRUE)
+
+pmean(..., na.rm = TRUE)
+
+pmedian(..., na.rm = TRUE)
+
+psd(..., na.rm = TRUE)
+
+pvar(..., na.rm = TRUE)
+
+pcv(..., na.rm = TRUE)
+
+pp01(..., na.rm = TRUE)
+
+pp025(..., na.rm = TRUE)
+
+pp05(..., na.rm = TRUE)
+
+pp10(..., na.rm = TRUE)
+
+pp25(..., na.rm = TRUE)
+
+pp75(..., na.rm = TRUE)
+
+pp95(..., na.rm = TRUE)
+
+pp975(..., na.rm = TRUE)
+
+pp99(..., na.rm = TRUE)Arguments
+- ... +
Numeric vectors of the same length
+
+
+- fun +
Function to apply
+
+
+- na.rm +
a logical indicating whether missing values should be removed.
+
+
Value
+ + +A vector of 'parallel' summaries of the argument vectors.
+Details
+Calculate summary statistics of the input vectors per row (or 'parallel')
+Examples
+pfun(1:10, fun = mean)
+#> [1] 1 2 3 4 5 6 7 8 9 10
+psum(1:10, 10:1)
+#> [1] 11 11 11 11 11 11 11 11 11 11
+Create a pivot table
+pivotr(
+ dataset,
+ cvars = "",
+ nvar = "None",
+ fun = "mean",
+ normalize = "None",
+ tabfilt = "",
+ tabsort = "",
+ tabslice = "",
+ nr = Inf,
+ data_filter = "",
+ arr = "",
+ rows = NULL,
+ envir = parent.frame()
+)Arguments
+- dataset +
Dataset to tabulate
+
+
+- cvars +
Categorical variables
+
+
+- nvar +
Numerical variable
+
+
+- fun +
Function to apply to numerical variable
+
+
+- normalize +
Normalize the table by row total, column totals, or overall total
+
+
+- tabfilt +
Expression used to filter the table (e.g., "Total > 10000")
+
+
+- tabsort +
Expression used to sort the table (e.g., "desc(Total)")
+
+
+- tabslice +
Expression used to filter table (e.g., "1:5")
+
+
+- nr +
Number of rows to display
+
+
+- data_filter +
Expression used to filter the dataset before creating the table (e.g., "price > 10000")
+
+
+- arr +
Expression to arrange (sort) the data on (e.g., "color, desc(price)")
+
+
+- rows +
Rows to select from the specified dataset
+
+
+- envir +
Environment to extract data from
+
+
Details
+Create a pivot-table. See https://radiant-rstats.github.io/docs/data/pivotr.html for an example in Radiant
+Examples
+pivotr(diamonds, cvars = "cut") %>% str()
+#> List of 18
+#> $ cni : logi [1:2] FALSE FALSE
+#> $ cn : chr [1:2] "cut" "n_obs"
+#> $ tab_freq : tibble [6 × 2] (S3: tbl_df/tbl/data.frame)
+#> ..$ cut : Factor w/ 6 levels "Fair","Good",..: 1 2 3 4 5 6
+#> ..$ n_obs: int [1:6] 101 275 677 771 1176 3000
+#> $ tab :'data.frame': 6 obs. of 2 variables:
+#> ..$ cut : Factor w/ 6 levels "Fair","Good",..: 1 2 3 4 5 6
+#> ..$ n_obs: int [1:6] 101 275 677 771 1176 3000
+#> ..- attr(*, "radiant_nrow")= num 5
+#> $ df_name : chr "diamonds"
+#> $ fill : int 0
+#> $ vars : chr "cut"
+#> $ cvars : chr "cut"
+#> $ nvar : chr "n_obs"
+#> $ fun : chr "mean"
+#> $ normalize : chr "None"
+#> $ tabfilt : chr ""
+#> $ tabsort : chr ""
+#> $ tabslice : chr ""
+#> $ nr : num Inf
+#> $ data_filter: chr ""
+#> $ arr : chr ""
+#> $ rows : NULL
+#> - attr(*, "class")= chr [1:2] "pivotr" "list"
+pivotr(diamonds, cvars = "cut")$tab
+#> cut n_obs
+#> 1 Fair 101
+#> 2 Good 275
+#> 3 Very Good 677
+#> 4 Premium 771
+#> 5 Ideal 1176
+#> 6 Total 3000
+pivotr(diamonds, cvars = c("cut", "clarity", "color"))$tab
+#> clarity color Fair Good Very_Good Premium Ideal Total
+#> 1 I1 D 0 1 0 3 0 4
+#> 2 I1 E 1 1 2 1 0 5
+#> 3 I1 F 2 1 2 2 4 11
+#> 4 I1 G 1 1 1 2 0 5
+#> 5 I1 H 3 0 1 3 1 8
+#> 6 I1 I 4 0 0 1 0 5
+#> 7 I1 J 1 0 0 1 0 2
+#> 8 SI2 D 8 14 18 13 15 68
+#> 9 SI2 E 5 16 30 25 30 106
+#> 10 SI2 F 1 11 24 23 34 93
+#> 11 SI2 G 5 7 29 35 21 97
+#> 12 SI2 H 4 7 14 31 30 86
+#> 13 SI2 I 2 2 7 21 14 46
+#> 14 SI2 J 3 5 10 9 6 33
+#> 15 SI1 D 4 9 21 38 39 111
+#> 16 SI1 E 5 22 37 47 36 147
+#> 17 SI1 F 4 14 40 25 42 125
+#> 18 SI1 G 5 10 23 26 31 95
+#> 19 SI1 H 9 13 21 31 43 117
+#> 20 SI1 I 1 14 18 20 23 76
+#> 21 SI1 J 1 11 11 9 18 50
+#> 22 VS2 D 2 3 17 12 55 89
+#> 23 VS2 E 2 15 26 41 55 139
+#> 24 VS2 F 4 12 23 36 53 128
+#> 25 VS2 G 2 8 17 42 50 119
+#> 26 VS2 H 2 10 20 25 31 88
+#> 27 VS2 I 2 2 16 17 19 56
+#> 28 VS2 J 0 2 12 12 16 42
+#> 29 VS1 D 0 3 10 13 24 50
+#> 30 VS1 E 0 5 9 13 25 52
+#> 31 VS1 F 4 8 25 17 29 83
+#> 32 VS1 G 2 7 22 30 51 112
+#> 33 VS1 H 3 3 19 23 22 70
+#> 34 VS1 I 2 5 11 14 18 50
+#> 35 VS1 J 2 2 3 10 8 25
+#> 36 VVS2 D 0 4 8 9 20 41
+#> 37 VVS2 E 1 1 25 7 22 56
+#> 38 VVS2 F 0 0 14 6 29 49
+#> 39 VVS2 G 1 3 21 10 43 78
+#> 40 VVS2 H 0 3 12 5 13 33
+#> 41 VVS2 I 0 1 1 4 14 20
+#> 42 VVS2 J 0 0 2 2 3 7
+#> 43 VVS1 D 1 1 2 4 9 17
+#> 44 VVS1 E 0 2 7 7 21 37
+#> 45 VVS1 F 1 7 4 4 29 45
+#> 46 VVS1 G 0 3 16 9 42 70
+#> 47 VVS1 H 0 1 6 11 14 32
+#> 48 VVS1 I 0 1 4 3 12 20
+#> 49 VVS1 J 0 0 1 1 1 3
+#> 50 IF D 0 0 1 0 1 2
+#> 51 IF E 0 0 4 3 5 12
+#> 52 IF F 1 2 4 6 18 31
+#> 53 IF G 0 1 2 2 16 21
+#> 54 IF H 0 0 2 3 15 20
+#> 55 IF I 0 1 2 3 5 11
+#> 56 IF J 0 0 0 1 1 2
+#> 57 Total Total 101 275 677 771 1176 3000
+pivotr(diamonds, cvars = "cut:clarity", nvar = "price")$tab
+#> clarity Fair Good Very_Good Premium Ideal Total
+#> 1 I1 2730.167 4333.500 3864.167 4932.231 6078.200 4194.775
+#> 2 SI2 5893.964 5280.919 5045.621 5568.019 4435.673 5100.189
+#> 3 SI1 4273.069 3757.022 4277.544 4113.811 3758.125 3998.577
+#> 4 VS2 3292.000 3925.481 3950.947 4522.914 3306.290 3822.967
+#> 5 VS1 5110.769 3740.697 3889.475 4461.333 3189.362 3789.181
+#> 6 VVS2 2030.500 4378.167 2525.193 3580.581 3665.181 3337.820
+#> 7 VVS1 6761.500 3889.333 1945.875 1426.692 2960.594 2608.460
+#> 8 IF 3205.000 817.250 4675.867 2361.333 1961.344 2411.697
+#> 9 Total 4505.238 4130.433 3959.916 4369.409 3470.224 3907.186
+pivotr(diamonds, cvars = "cut", nvar = "price")$tab
+#> cut price
+#> 1 Fair 4505.238
+#> 2 Good 4130.433
+#> 3 Very Good 3959.916
+#> 4 Premium 4369.409
+#> 5 Ideal 3470.224
+#> 6 Total 3907.186
+pivotr(diamonds, cvars = "cut", normalize = "total")$tab
+#> cut n_obs
+#> 1 Fair 0.03366667
+#> 2 Good 0.09166667
+#> 3 Very Good 0.22566667
+#> 4 Premium 0.25700000
+#> 5 Ideal 0.39200000
+#> 6 Total 1.00000000
+
+Plot method for the pivotr function
+# S3 method for pivotr
+plot(
+ x,
+ type = "dodge",
+ perc = FALSE,
+ flip = FALSE,
+ fillcol = "blue",
+ opacity = 0.5,
+ ...
+)Arguments
+- x +
Return value from
pivotr
+
+
+- type +
Plot type to use ("fill" or "dodge" (default))
+
+
+- perc +
Use percentage on the y-axis
+
+
+- flip +
Flip the axes in a plot (FALSE or TRUE)
+
+
+- fillcol +
Fill color for bar-plot when only one categorical variable has been selected (default is "blue")
+
+
+- opacity +
Opacity for plot elements (0 to 1)
+
+
+- ... +
further arguments passed to or from other methods
+
+
Details
+See https://radiant-rstats.github.io/docs/data/pivotr for an example in Radiant
+See also
+pivotr to generate summaries
summary.pivotr to show summaries
Print/draw method for grobs produced by gridExtra
+# S3 method for gtable +print(x, ...)+ +
Arguments
+| x | +a gtable object |
+
|---|---|
| ... | +further arguments passed to or from other methods |
+
Value
+ +A plot
+Details
+ +Print method for grobs created using grid.arrange
+ +Calculate proportion
+prop(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Proportion of first level for a factor and of the maximum value for numeric
+Comic publishers
+data(publishers)Format
+A data frame with 3 rows and 2 variables
+Details
+List of comic publishers from https://stat545.com/join-cheatsheet.html. The dataset is used to illustrate data merging / joining. Description provided in attr(publishers,"description")
+Create a qscatter plot similar to Stata
+qscatter(dataset, xvar, yvar, lev = "", fun = "mean", bins = 20)Arguments
+- dataset +
Data to plot (data.frame or tibble)
+
+
+- xvar +
Character indicating the variable to display along the X-axis of the plot
+
+
+- yvar +
Character indicating the variable to display along the Y-axis of the plot
+
+
+- lev +
Level in yvar to use if yvar is of type character of factor. If lev is empty then the first level is used
+
+
+- fun +
Summary measure to apply to both the x and y variable
+
+
+- bins +
Number of bins to use
+
+
Examples
+qscatter(diamonds, "price", "carat")
+ +qscatter(titanic, "age", "survived")
+
+qscatter(titanic, "age", "survived")
+ +
+
+
+Create a vector of quadratic and cubed terms for use in linear and logistic regression
+ Source:R/radiant.R
+ qterms.RdCreate a vector of quadratic and cubed terms for use in linear and logistic regression
+qterms(vars, nway = 2)Arguments
+- vars +
Variables labels to use
+
+
+- nway +
quadratic (2) or cubic (3) term labels to create
+
+
Value
+ + +Character vector of (regression) term labels
+Deprecated function(s) in the radiant.data package
+ Source:R/deprecated.R
+ radiant.data-deprecated.RdThese functions are provided for compatibility with previous versions of +radiant but will be removed
+mean_rm(...)Arguments
+- ... +
Parameters to be passed to the updated functions
+
+
Details
+ + +Replace
mean_rmbymean
+Replace
median_rmbymedian
+Replace
min_rmbymin
+Replace
max_rmbymax
+Replace
sd_rmbysd
+Replace
var_rmbyvar
+Replace
sum_rmbysum
+Replace
getdatabyget_data
+Replace
filterdatabyfilter_data
+Replace
combinedatabycombine_data
+Replace
viewdatabyview_data
+Replace
toFctbyto_fct
+Replace
fixMSbyfix_smart
+Replace
rounddfbyround_df
+Replace
formatdfbyformat_df
+Replace
formatnrbyformat_nr
+Replace
getclassbyget_class
+Replace
is_numericbyis_double
+Replace
is_emptybyis.empty
+
Launch the radiant.data app in the default web browser
+radiant.data(state, ...)Arguments
+- state +
Path to statefile to load
+
+
+- ... +
additional arguments to pass to shiny::runApp (e.g, port = 8080)
+
+
Examples
+if (FALSE) {
+radiant.data()
+radiant.data("https://github.com/radiant-rstats/docs/raw/gh-pages/examples/demo-dvd-rnd.state.rda")
+radiant.data("viewer")
+}
+Start radiant.data app but do not open a browser
+radiant.data_url(state, ...)Arguments
+- state +
Path to statefile to load
+
+
+- ... +
additional arguments to pass to shiny::runApp (e.g, port = 8080)
+
+
Examples
+if (FALSE) {
+radiant.data_url()
+}
+Launch the radiant.data app in the Rstudio viewer
+ Source:R/radiant.R
+ radiant.data_viewer.RdLaunch the radiant.data app in the Rstudio viewer
+radiant.data_viewer(state, ...)Arguments
+- state +
Path to statefile to load
+
+
+- ... +
additional arguments to pass to shiny::runApp (e.g, port = 8080)
+
+
Examples
+if (FALSE) {
+radiant.data_viewer()
+}
+Launch the radiant.data app in an Rstudio window
+radiant.data_window(state, ...)Arguments
+- state +
Path to statefile to load
+
+
+- ... +
additional arguments to pass to shiny::runApp (e.g, port = 8080)
+
+
Examples
+if (FALSE) {
+radiant.data_window()
+}
+Generate code to read a file
+read_files(
+ path,
+ pdir = "",
+ type = "rmd",
+ to = "",
+ clipboard = TRUE,
+ radiant = FALSE
+)Arguments
+- path +
Path to file. If empty, a file browser will be opened
+
+
+- pdir +
Project dir
+
+
+- type +
Generate code for _Report > Rmd_ ("rmd") or _Report > R_ ("r")
+
+
+- to +
Name to use for object. If empty, will use file name to derive an object name
+
+
+- clipboard +
Return code to clipboard (not available on Linux)
+
+
+- radiant +
Should returned code be formatted for use with other code generated by Radiant?
+
+
Details
+Return code to read a file at the specified path. Will open a file browser if no path is provided
+Examples
+if (interactive()) {
+ read_files(clipboard = FALSE)
+}
+These objects are imported from other packages. Follow the links +below to see their documentation.
+- broom +
- + + +
- bslib +
- + + +
- glue +
- + + +
- knitr +
- + + +
- lubridate +
- + + +
- patchwork +
- + + +
- png +
- + + +
- psych +
- + + +
- tibble +
- + + +
Remove/reorder levels
+refactor(x, levs = levels(x), repl = NA)Arguments
+- x +
Character or Factor
+
+
+- levs +
Set of levels to use
+
+
+- repl +
String (or NA) used to replace missing levels
+
+
Details
+Keep only a specific set of levels in a factor. By removing levels the base for comparison in, e.g., regression analysis, becomes the first level. To relabel the base use, for example, repl = 'other'
+Register a data.frame or list in Radiant
+register(
+ new,
+ org = "",
+ descr = "",
+ shiny = shiny::getDefaultReactiveDomain(),
+ envir = r_data
+)Arguments
+- new +
String containing the name of the data.frame to register
+
+
+- org +
Name of the original data.frame if a (working) copy is being made
+
+
+- descr +
Data description in markdown format
+
+
+- shiny +
Check if function is called from a shiny application
+
+
+- envir +
Environment to assign data to
+
+
See also
+See also add_description to add a description in markdown format
+ to a data.frame
Method to render DT tables
+# S3 method for datatables
+render(object, shiny = shiny::getDefaultReactiveDomain(), ...)Arguments
+- object +
DT table
+
+
+- shiny +
Check if function is called from a shiny application
+
+
+- ... +
Additional arguments
+
+
Base method used to render htmlwidgets
+render(object, ...)Arguments
+- object +
Object of relevant class to render
+
+
+- ... +
Additional arguments
+
+
Method to render plotly plots
+# S3 method for plotly
+render(object, shiny = shiny::getDefaultReactiveDomain(), ...)Arguments
+- object +
plotly object
+
+
+- shiny +
Check if function is called from a shiny application
+
+
+- ... +
Additional arguments
+
+
Round doubles in a data.frame to a specified number of decimal places
+ Source:R/radiant.R
+ round_df.RdRound doubles in a data.frame to a specified number of decimal places
+round_df(tbl, dec = 3)Arguments
+- tbl +
Data frame
+
+
+- dec +
Number of decimals to show
+
+
Value
+ + +Data frame with rounded doubles
+Examples
+data.frame(x = as.factor(c("a", "b")), y = c(1L, 2L), z = c(-0.0005, 3.1)) %>%
+ round_df(dec = 2)
+#> x y z
+#> 1 a 1 0.0
+#> 2 b 2 3.1
+Save data to clipboard on Windows or macOS
+save_clip(dataset)Arguments
+- dataset +
Dataset to save to clipboard
+
+
Details
+Save a data.frame or tibble to the clipboard on Windows or macOS
+See also
+See the load_clip
Standard deviation for the population
+sdpop(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Standard deviation for the population
+Examples
+sdpop(rnorm(100))
+#> [1] 0.9326249
+
+Standard deviation for proportion
+sdprop(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Standard deviation for proportion
+Standard error
+se(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Standard error
+Examples
+se(rnorm(100))
+#> [1] 0.1073877
+
+Search for a pattern in all columns of a data.frame
+search_data(dataset, pattern, ignore.case = TRUE, fixed = FALSE)Arguments
+- dataset +
Data.frame to search
+
+
+- pattern +
String to match
+
+
+- ignore.case +
Should search be case sensitive or not (default is FALSE)
+
+
+- fixed +
Allow regular expressions or not (default is FALSE)
+
+
See also
+See grepl for a detailed description of the function arguments
Examples
+publishers %>% filter(search_data(., "^m"))
+#> # A tibble: 1 × 2
+#> publisher yr_founded
+#> <chr> <int>
+#> 1 Marvel 1939
+Standard error for proportion
+seprop(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Standard error for proportion
+Alias used to add an attribute
+set_attr(x, which, value)Arguments
+- x +
Object
+
+
+- which +
Attribute name
+
+
+- value +
Value to set
+
+
Examples
+foo <- data.frame(price = 1:5) %>% set_attr("description", "price set in experiment ...")
+Show all rows with duplicated values (not just the first or last)
+ Source:R/transform.R
+ show_duplicated.RdShow all rows with duplicated values (not just the first or last)
+show_duplicated(.tbl, ...)Arguments
+- .tbl +
Data frame to add transformed variables to
+
+
+- ... +
Variables used to evaluate row uniqueness
+
+
Details
+If an entire row is duplicated use "duplicated" to show only one of the duplicated rows. When using a subset of variables to establish uniqueness it may be of interest to show all rows that have (some) duplicate elements
+Examples
+bind_rows(mtcars, mtcars[c(1, 5, 7), ]) %>%
+ show_duplicated(mpg, cyl)
+#> # A tibble: 15 × 12
+#> mpg cyl disp hp drat wt qsec vs am gear carb nr_dup
+#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
+#> 1 10.4 8 472 205 2.93 5.25 18.0 0 0 3 4 1
+#> 2 10.4 8 460 215 3 5.42 17.8 0 0 3 4 2
+#> 3 14.3 8 360 245 3.21 3.57 15.8 0 0 3 4 1
+#> 4 14.3 8 360 245 3.21 3.57 15.8 0 0 3 4 2
+#> 5 15.2 8 276. 180 3.07 3.78 18 0 0 3 3 1
+#> 6 15.2 8 304 150 3.15 3.44 17.3 0 0 3 2 2
+#> 7 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2 1
+#> 8 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2 2
+#> 9 21 6 160 110 3.9 2.62 16.5 0 1 4 4 1
+#> 10 21 6 160 110 3.9 2.88 17.0 0 1 4 4 2
+#> 11 21 6 160 110 3.9 2.62 16.5 0 1 4 4 3
+#> 12 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1 1
+#> 13 22.8 4 141. 95 3.92 3.15 22.9 1 0 4 2 2
+#> 14 30.4 4 75.7 52 4.93 1.62 18.5 1 1 4 2 1
+#> 15 30.4 4 95.1 113 3.77 1.51 16.9 1 1 5 2 2
+bind_rows(mtcars, mtcars[c(1, 5, 7), ]) %>%
+ show_duplicated()
+#> mpg cyl disp hp drat wt qsec vs am gear carb
+#> Mazda RX4 21.0 6 160 110 3.90 2.62 16.46 0 1 4 4
+#> Hornet Sportabout 18.7 8 360 175 3.15 3.44 17.02 0 0 3 2
+#> Duster 360 14.3 8 360 245 3.21 3.57 15.84 0 0 3 4
+
+Add stars based on p.values
+sig_stars(pval)Arguments
+- pval +
Vector of p-values
+
+
Value
+ + +A vector of stars
+Examples
+sig_stars(c(.0009, .049, .009, .4, .09))
+#> [1] "***" "*" "**" "" "."
+Slice data with user-specified expression
+slice_data(dataset, expr = NULL, drop = TRUE)Arguments
+- dataset +
Data frame to slice
+
+
+- expr +
Expression to use select rows from the specified dataset
+
+
+- drop +
Drop unused factor levels after filtering (default is TRUE)
+
+
Value
+ + +Sliced data frame
+Details
+Select only a slice of the data to work with
+Calculate square of a variable
+square(x)Arguments
+- x +
Input variable
+
+
Value
+ + +x^2
+Hide warnings and messages and return invisible
+sshh(...)Arguments
+- ... +
Inputs to keep quite
+
+
Details
+Hide warnings and messages and return invisible
+Hide warnings and messages and return result
+sshhr(...)Arguments
+- ... +
Inputs to keep quite
+
+
Details
+Hide warnings and messages and return result
+Standardize
+standardize(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +If x is a numeric variable return (x - mean(x)) / sd(x)
+Deprecated: Store method for the explore function
+# S3 method for explore
+store(dataset, object, name, ...)Arguments
+- dataset +
Dataset
+
+
+- object +
Return value from
explore
+
+
+- name +
Name to assign to the dataset
+
+
+- ... +
further arguments passed to or from other methods
+
+
Details
+Return the summarized data. See https://radiant-rstats.github.io/docs/data/explore.html for an example in Radiant
+See also
+explore to generate summaries
Method to store variables in a dataset in Radiant
+store(dataset, object = "deprecated", ...)Arguments
+- dataset +
Dataset
+
+
+- object +
Object of relevant class that has information to be stored
+
+
+- ... +
Additional arguments
+
+
Deprecated: Store method for the pivotr function
+# S3 method for pivotr
+store(dataset, object, name, ...)Arguments
+- dataset +
Dataset
+
+
+- object +
Return value from
pivotr
+
+
+- name +
Name to assign to the dataset
+
+
+- ... +
further arguments passed to or from other methods
+
+
Details
+Return the summarized data. See https://radiant-rstats.github.io/docs/data/pivotr.html for an example in Radiant
+See also
+pivotr to generate summaries
Work around to avoid (harmless) messages from subplot
+subplot(..., margin = 0.04)Arguments
+- ... +
Arguments to pass to the
subplotfunction in the plotly packages
+
+
+- margin +
Default margin to use between plots
+
+
See also
+See the subplot in the plotly package for details (?plotly::subplot)
Summary method for the explore function
+# S3 method for explore
+summary(object, dec = 3, ...)Arguments
+- object +
Return value from
explore
+
+
+- dec +
Number of decimals to show
+
+
+- ... +
further arguments passed to or from other methods
+
+
Details
+See https://radiant-rstats.github.io/docs/data/explore.html for an example in Radiant
+See also
+explore to generate summaries
Examples
+result <- explore(diamonds, "price:x")
+summary(result)
+#> Explore
+#> Data : diamonds
+#> Functions : mean, sd
+#> Top : Function
+#>
+#> variable mean sd
+#> price 3,907.186 3,956.915
+#> carat 0.794 0.474
+#> clarity 0.013 0.115
+#> cut 0.034 0.180
+#> color 0.127 0.333
+#> depth 61.753 1.446
+#> table 57.465 2.241
+#> x 5.722 1.124
+result <- explore(diamonds, "price", byvar = "cut", fun = c("n_obs", "skew"))
+summary(result)
+#> Explore
+#> Data : diamonds
+#> Grouped by : cut
+#> Functions : n_obs, skew
+#> Top : Function
+#>
+#> cut variable n_obs skew
+#> Fair price 101 1.574
+#> Good price 275 1.489
+#> Very Good price 677 1.601
+#> Premium price 771 1.413
+#> Ideal price 1,176 1.799
+explore(diamonds, "price:x", byvar = "color") %>% summary()
+#> Explore
+#> Data : diamonds
+#> Grouped by : color
+#> Functions : mean, sd
+#> Top : Function
+#>
+#> color variable mean sd
+#> D price 3,217.003 3,278.276
+#> D carat 0.665 0.365
+#> D clarity 0.010 0.102
+#> D cut 0.039 0.194
+#> D depth 61.705 1.452
+#> D table 57.438 2.269
+#> D x 5.437 0.943
+#> E price 3,284.596 3,610.872
+#> E carat 0.679 0.391
+#> E clarity 0.009 0.095
+#> E cut 0.025 0.157
+#> E depth 61.768 1.391
+#> E table 57.548 2.262
+#> E x 5.455 0.999
+#> F price 3,654.492 3,779.511
+#> F carat 0.728 0.405
+#> F clarity 0.019 0.138
+#> F cut 0.030 0.171
+#> F depth 61.686 1.396
+#> F table 57.428 2.275
+#> F x 5.590 1.016
+#> G price 3,970.573 4,002.082
+#> G carat 0.774 0.451
+#> G clarity 0.008 0.091
+#> G cut 0.027 0.162
+#> G depth 61.669 1.404
+#> G table 57.341 2.133
+#> G x 5.686 1.094
+#> H price 4,250.302 4,063.648
+#> H carat 0.880 0.503
+#> H clarity 0.018 0.132
+#> H cut 0.046 0.210
+#> H depth 61.841 1.463
+#> H table 57.464 2.208
+#> H x 5.914 1.186
+#> I price 4,869.190 4,570.572
+#> I carat 1.001 0.573
+#> I clarity 0.018 0.132
+#> I cut 0.039 0.193
+#> I depth 61.938 1.620
+#> I table 57.446 2.160
+#> I x 6.163 1.251
+#> J price 5,642.012 4,574.576
+#> J carat 1.193 0.602
+#> J clarity 0.012 0.110
+#> J cut 0.043 0.203
+#> J depth 61.779 1.549
+#> J table 57.863 2.563
+#> J x 6.578 1.237
+
+Summary method for pivotr
+# S3 method for pivotr
+summary(object, perc = FALSE, dec = 3, chi2 = FALSE, shiny = FALSE, ...)Arguments
+- object +
Return value from
pivotr
+
+
+- perc +
Display numbers as percentages (TRUE or FALSE)
+
+
+- dec +
Number of decimals to show
+
+
+- chi2 +
If TRUE calculate the chi-square statistic for the (pivot) table
+
+
+- shiny +
Did the function call originate inside a shiny app
+
+
+- ... +
further arguments passed to or from other methods
+
+
Details
+See https://radiant-rstats.github.io/docs/data/pivotr.html for an example in Radiant
+See also
+pivotr to create the pivot-table using dplyr
Examples
+pivotr(diamonds, cvars = "cut") %>% summary(chi2 = TRUE)
+#> Pivot table
+#> Data : diamonds
+#> Categorical : cut
+#>
+#> cut n_obs
+#> Fair 101
+#> Good 275
+#> Very Good 677
+#> Premium 771
+#> Ideal 1,176
+#> Total 3,000
+#>
+#> Chi-squared: 1202.62 df(4), p.value < .001
+#> 0.0% of cells have expected values below 5
+pivotr(diamonds, cvars = "cut", tabsort = "desc(n_obs)") %>% summary()
+#> Pivot table
+#> Data : diamonds
+#> Table sorted: desc(n_obs)
+#> Categorical : cut
+#>
+#> cut n_obs
+#> Ideal 1,176
+#> Premium 771
+#> Very Good 677
+#> Good 275
+#> Fair 101
+#> Total 3,000
+#>
+pivotr(diamonds, cvars = "cut", tabfilt = "n_obs > 700") %>% summary()
+#> Pivot table
+#> Data : diamonds
+#> Table filter: n_obs > 700
+#> Categorical : cut
+#>
+#> cut n_obs
+#> Premium 771
+#> Ideal 1,176
+#> Total 3,000
+#>
+pivotr(diamonds, cvars = "cut:clarity", nvar = "price") %>% summary()
+#> Pivot table
+#> Data : diamonds
+#> Categorical : cut clarity
+#> Numeric : price
+#> Function : mean
+#>
+#> clarity Fair Good Very_Good Premium Ideal Total
+#> I1 2,730.167 4,333.500 3,864.167 4,932.231 6,078.200 4,194.775
+#> SI2 5,893.964 5,280.919 5,045.621 5,568.019 4,435.673 5,100.189
+#> SI1 4,273.069 3,757.022 4,277.544 4,113.811 3,758.125 3,998.577
+#> VS2 3,292.000 3,925.481 3,950.947 4,522.914 3,306.290 3,822.967
+#> VS1 5,110.769 3,740.697 3,889.475 4,461.333 3,189.362 3,789.181
+#> VVS2 2,030.500 4,378.167 2,525.193 3,580.581 3,665.181 3,337.820
+#> VVS1 6,761.500 3,889.333 1,945.875 1,426.692 2,960.594 2,608.460
+#> IF 3,205.000 817.250 4,675.867 2,361.333 1,961.344 2,411.697
+#> Total 4,505.238 4,130.433 3,959.916 4,369.409 3,470.224 3,907.186
+#>
+
+Super heroes
+data(superheroes)Format
+A data frame with 7 rows and 4 variables
+Details
+List of super heroes from https://stat545.com/join-cheatsheet.html. The dataset is used to illustrate data merging / joining. Description provided in attr(superheroes,"description")
+Create data.frame from a table
+Arguments
+- dataset +
Data.frame
+
+
+- freq +
Column name with frequency information
+
+
Examples
+data.frame(price = c("$200", "$300"), sale = c(10, 2)) %>% table2data()
+#> price
+#> 1 $200
+#> 1.1 $200
+#> 1.2 $200
+#> 1.3 $200
+#> 1.4 $200
+#> 1.5 $200
+#> 1.6 $200
+#> 1.7 $200
+#> 1.8 $200
+#> 1.9 $200
+#> 2 $300
+#> 2.1 $300
+
+Convert characters to factors
+to_fct(dataset, safx = 30, nuniq = 100, n = 100)Arguments
+- dataset +
Data frame
+
+
+- safx +
Ratio of number of rows to number of unique values
+
+
+- nuniq +
Cutoff for number of unique values
+
+
+- n +
Cutoff for small dataset
+
+
Details
+Convert columns of type character to factors based on a set of rules. By default columns will be converted for small datasets (<= 100 rows) with more rows than unique values. For larger datasets, columns are converted only when the number of unique values is <= 100 and there are 30 or more rows in the data for every unique value
+Variance for the population
+varpop(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Variance for the population
+Examples
+varpop(rnorm(100))
+#> [1] 1.184664
+
+Variance for proportion
+varprop(x, na.rm = TRUE)Arguments
+- x +
Input variable
+
+
+- na.rm +
If TRUE missing values are removed before calculation
+
+
Value
+ + +Variance for proportion
+View data in a shiny-app
+view_data(
+ dataset,
+ vars = "",
+ filt = "",
+ arr = "",
+ rows = NULL,
+ na.rm = FALSE,
+ dec = 3,
+ envir = parent.frame()
+)Arguments
+- dataset +
Data.frame or name of the dataframe to view
+
+
+- vars +
Variables to show (default is all)
+
+
+- filt +
Filter to apply to the specified dataset
+
+
+- arr +
Expression to arrange (sort) data
+
+
+- rows +
Select rows in the specified dataset
+
+
+- na.rm +
Remove rows with missing values (default is FALSE)
+
+
+- dec +
Number of decimals to show
+
+
+- envir +
Environment to extract data from
+
+
Details
+View, search, sort, etc. your data
+See also
+See get_data and filter_data
Examples
+if (FALSE) {
+view_data(mtcars)
+}
+
+Visualize data using ggplot2 https://ggplot2.tidyverse.org/
+ Source:R/visualize.R
+ visualize.RdVisualize data using ggplot2 https://ggplot2.tidyverse.org/
+visualize(
+ dataset,
+ xvar,
+ yvar = "",
+ comby = FALSE,
+ combx = FALSE,
+ type = ifelse(is.empty(yvar), "dist", "scatter"),
+ nrobs = -1,
+ facet_row = ".",
+ facet_col = ".",
+ color = "none",
+ fill = "none",
+ size = "none",
+ fillcol = "blue",
+ linecol = "black",
+ pointcol = "black",
+ bins = 10,
+ smooth = 1,
+ fun = "mean",
+ check = "",
+ axes = "",
+ alpha = 0.5,
+ theme = "theme_gray",
+ base_size = 11,
+ base_family = "",
+ labs = list(),
+ xlim = NULL,
+ ylim = NULL,
+ data_filter = "",
+ arr = "",
+ rows = NULL,
+ shiny = FALSE,
+ custom = FALSE,
+ envir = parent.frame()
+)Arguments
+- dataset +
Data to plot (data.frame or tibble)
+
+
+- xvar +
One or more variables to display along the X-axis of the plot
+
+
+- yvar +
Variable to display along the Y-axis of the plot (default = "none")
+
+
+- comby +
Combine yvars in plot (TRUE or FALSE, FALSE is the default)
+
+
+- combx +
Combine xvars in plot (TRUE or FALSE, FALSE is the default)
+
+
+- type +
Type of plot to create. One of Distribution ('dist'), Density ('density'), Scatter ('scatter'), Surface ('surface'), Line ('line'), Bar ('bar'), or Box-plot ('box')
+
+
+- nrobs +
Number of data points to show in scatter plots (-1 for all)
+
+
+- facet_row +
Create vertically arranged subplots for each level of the selected factor variable
+
+
+- facet_col +
Create horizontally arranged subplots for each level of the selected factor variable
+
+
+- color +
Adds color to a scatter plot to generate a 'heat map'. For a line plot one line is created for each group and each is assigned a different color
+
+
+- fill +
Display bar, distribution, and density plots by group, each with a different color. Also applied to surface plots to generate a 'heat map'
+
+
+- size +
Numeric variable used to scale the size of scatter-plot points
+
+
+- fillcol +
Color used for bars, boxes, etc. when no color or fill variable is specified
+
+
+- linecol +
Color for lines when no color variable is specified
+
+
+- pointcol +
Color for points when no color variable is specified
+
+
+- bins +
Number of bins used for a histogram (1 - 50)
+
+
+- smooth +
Adjust the flexibility of the loess line for scatter plots
+
+
+- fun +
Set the summary measure for line and bar plots when the X-variable is a factor (default is "mean"). Also used to plot an error bar in a scatter plot when the X-variable is a factor. Options are "mean" and/or "median"
+
+
+- check +
Add a regression line ("line"), a loess line ("loess"), or jitter ("jitter") to a scatter plot
+
+
+- axes +
Flip the axes in a plot ("flip") or apply a log transformation (base e) to the y-axis ("log_y") or the x-axis ("log_x")
+
+
+- alpha +
Opacity for plot elements (0 to 1)
+
+
+- theme +
ggplot theme to use (e.g., "theme_gray" or "theme_classic")
+
+
+- base_size +
Base font size to use (default = 11)
+
+
+- base_family +
Base font family to use (e.g., "Times" or "Helvetica")
+
+
+- labs +
Labels to use for plots
+
+
+- xlim +
Set limit for x-axis (e.g., c(0, 1))
+
+
+- ylim +
Set limit for y-axis (e.g., c(0, 1))
+
+
+- data_filter +
Expression used to filter the dataset. This should be a string (e.g., "price > 10000")
+
+
+- arr +
Expression used to sort the data. Likely used in combination for `rows`
+
+
+- rows +
Rows to select from the specified dataset
+
+
+- shiny +
Logical (TRUE, FALSE) to indicate if the function call originate inside a shiny app
+
+
+- custom +
Logical (TRUE, FALSE) to indicate if ggplot object (or list of ggplot objects) should be returned. This option can be used to customize plots (e.g., add a title, change x and y labels, etc.). See examples and https://ggplot2.tidyverse.org for options.
+
+
+- envir +
Environment to extract data from
+
+
Value
+ + +Generated plots
+Details
+See https://radiant-rstats.github.io/docs/data/visualize.html for an example in Radiant
+Examples
+visualize(diamonds, "price:cut", type = "dist", fillcol = "red")
+ +visualize(diamonds, "carat:cut",
+ yvar = "price", type = "scatter",
+ pointcol = "blue", fun = c("mean", "median"), linecol = c("red", "green")
+)
+
+visualize(diamonds, "carat:cut",
+ yvar = "price", type = "scatter",
+ pointcol = "blue", fun = c("mean", "median"), linecol = c("red", "green")
+)
+ +visualize(diamonds,
+ yvar = "price", xvar = c("cut", "clarity"),
+ type = "bar", fun = "median"
+)
+
+visualize(diamonds,
+ yvar = "price", xvar = c("cut", "clarity"),
+ type = "bar", fun = "median"
+)
+ +visualize(diamonds,
+ yvar = "price", xvar = c("cut", "clarity"),
+ type = "line", fun = "max"
+)
+
+visualize(diamonds,
+ yvar = "price", xvar = c("cut", "clarity"),
+ type = "line", fun = "max"
+)
+ +visualize(diamonds,
+ yvar = "price", xvar = "carat", type = "scatter",
+ size = "table", custom = TRUE
+) + scale_size(range = c(1, 10), guide = "none")
+
+visualize(diamonds,
+ yvar = "price", xvar = "carat", type = "scatter",
+ size = "table", custom = TRUE
+) + scale_size(range = c(1, 10), guide = "none")
+ +visualize(diamonds, yvar = "price", xvar = "carat", type = "scatter", custom = TRUE) +
+ labs(title = "A scatterplot", x = "price in $")
+
+visualize(diamonds, yvar = "price", xvar = "carat", type = "scatter", custom = TRUE) +
+ labs(title = "A scatterplot", x = "price in $")
+ +visualize(diamonds, xvar = "price:carat", custom = TRUE) %>%
+ wrap_plots(ncol = 2) + plot_annotation(title = "Histograms")
+
+visualize(diamonds, xvar = "price:carat", custom = TRUE) %>%
+ wrap_plots(ncol = 2) + plot_annotation(title = "Histograms")
+ +visualize(diamonds,
+ xvar = "cut", yvar = "price", type = "bar",
+ facet_row = "cut", fill = "cut"
+)
+
+visualize(diamonds,
+ xvar = "cut", yvar = "price", type = "bar",
+ facet_row = "cut", fill = "cut"
+)
+ +
+
+
+Add ordered argument to lubridate::wday
+wday(x, label = FALSE, abbr = TRUE, ordered = FALSE)Arguments
+- x +
Input date vector
+
+
+- label +
Weekday as label (TRUE, FALSE)
+
+
+- abbr +
Abbreviate label (TRUE, FALSE)
+
+
+- ordered +
Order factor (TRUE, FALSE)
+
+
See also
+See the lubridate::wday() function in the lubridate package for additional details
Weighted standard deviation
+weighted.sd(x, wt, na.rm = TRUE)Arguments
+- x +
Numeric vector
+
+
+- wt +
Numeric vector of weights
+
+
+- na.rm +
Remove missing values (default is TRUE)
+
+
Details
+Calculate weighted standard deviation
+Index of the maximum per row
+which.pmax(...)Arguments
+- ... +
Numeric or character vectors of the same length
+
+
Value
+ + +Vector of rankings
+Details
+Determine the index of the maximum of the input vectors per row. Extension of which.max
See also
+See also which.max and which.pmin
Examples
+which.pmax(1:10, 10:1)
+#> [1] 2 2 2 2 2 1 1 1 1 1
+which.pmax(2, 10:1)
+#> [1] 2 2 2 2 2 2 2 2 1 1
+which.pmax(mtcars)
+#> [1] 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 4 3 4 4 3
+Index of the minimum per row
+which.pmin(...)Arguments
+- ... +
Numeric or character vectors of the same length
+
+
Value
+ + +Vector of rankings
+Details
+Determine the index of the minimum of the input vectors per row. Extension of which.min
See also
+See also which.min and which.pmax
Examples
+which.pmin(1:10, 10:1)
+#> [1] 1 1 1 1 1 2 2 2 2 2
+which.pmin(2, 10:1)
+#> [1] 1 1 1 1 1 1 1 1 1 2
+which.pmin(mtcars)
+#> [1] 8 8 8 9 8 9 8 9 9 9 9 8 8 8 8 8 8 8 8 8 9 8 8 8 8 8 8 8 8 8 8 8
+Workaround to store description file together with a parquet data file
+ Source:R/radiant.R
+ write_parquet.RdWorkaround to store description file together with a parquet data file
+write_parquet(x, file, description = attr(x, "description"))Arguments
+- x +
A data frame to write to disk
+
+
+- file +
Path to store parquet file
+
+
+- description +
Data description
+
+
Split a numeric variable into a number of bins and return a vector of bin numbers
+ Source:R/transform.R
+ xtile.RdSplit a numeric variable into a number of bins and return a vector of bin numbers
+xtile(x, n = 5, rev = FALSE, type = 7)Arguments
+- x +
Numeric variable
+
+
+- n +
number of bins to create
+
+
+- rev +
Reverse the order of the bin numbers
+
+
+- type +
An integer between 1 and 9 to select one of the quantile algorithms described in the help for the stats::quantile function
+
+
See also
+See quantile for a description of the different algorithm types
 ")
+options(radiant.help.cc.sa = "© Vincent Nijs (2023)
")
+options(radiant.help.cc.sa = "© Vincent Nijs (2023)  ")
+
+#####################################
+## url processing to share results
+#####################################
+
+## relevant links
+# http://stackoverflow.com/questions/25306519/shiny-saving-url-state-subpages-and-tabs/25385474#25385474
+# https://groups.google.com/forum/#!topic/shiny-discuss/Xgxq08N8HBE
+# https://gist.github.com/jcheng5/5427d6f264408abf3049
+
+## try http://127.0.0.1:3174/?url=multivariate/conjoint/plot/&SSUID=local
+options(
+ radiant.url.list =
+ list("Data" = list("tabs_data" = list(
+ "Manage" = "data/",
+ "View" = "data/view/",
+ "Visualize" = "data/visualize/",
+ "Pivot" = "data/pivot/",
+ "Explore" = "data/explore/",
+ "Transform" = "data/transform/",
+ "Combine" = "data/combine/",
+ "Rmd" = "report/rmd/",
+ "R" = "report/r/"
+ )))
+)
+
+make_url_patterns <- function(url_list = getOption("radiant.url.list"),
+ url_patterns = list()) {
+ for (i in names(url_list)) {
+ res <- url_list[[i]]
+ if (!is.list(res)) {
+ url_patterns[[res]] <- list("nav_radiant" = i)
+ } else {
+ tabs <- names(res)
+ for (j in names(res[[tabs]])) {
+ url <- res[[tabs]][[j]]
+ url_patterns[[url]] <- setNames(list(i, j), c("nav_radiant", tabs))
+ }
+ }
+ }
+ url_patterns
+}
+
+## generate url patterns
+options(radiant.url.patterns = make_url_patterns())
+
+## installed packages versions
+tmp <- grep("radiant\\.", installed.packages()[, "Package"], value = TRUE)
+if ("radiant" %in% installed.packages()) {
+ tmp <- c("radiant" = "radiant", tmp)
+}
+
+if (length(tmp) > 0) {
+ radiant.versions <- sapply(names(tmp), function(x) paste(x, paste(packageVersion(x), sep = "."))) %>% unique()
+ if ("shiny" %in% installed.packages()) {
+ radiant.versions <- c(radiant.versions, paste("shiny ", packageVersion("shiny")))
+ }
+} else {
+ radiant.versions <- "Unknown"
+}
+
+options(radiant.versions = paste(radiant.versions, collapse = ", "))
+rm(tmp, radiant.versions)
+
+if (is.null(getOption("radiant.theme", default = NULL))) {
+ options(radiant.theme = bslib::bs_theme(version = 4))
+}
+
+## bslib theme and version
+has_bslib_theme <- function() {
+ if (rlang::is_installed("bslib")) bslib::is_bs_theme(getOption("radiant.theme")) else FALSE
+}
+
+bslib_current_version <- function() {
+ if (rlang::is_installed("bslib")) bslib::theme_version(getOption("radiant.theme", default = bslib::bs_theme(version = 4)))
+}
+
+navbar_proj <- function(navbar) {
+ pdir <- radiant.data::find_project(mess = FALSE)
+ if (radiant.data::is.empty(pdir)) {
+ if (getOption("radiant.shinyFiles", FALSE) && !radiant.data::is.empty(getOption("radiant.sf_volumes", ""))) {
+ proj <- paste0(i18n$t("Base dir: "), names(getOption("radiant.sf_volumes"))[1])
+ } else {
+ proj <- "Project: (None)"
+ }
+ options(radiant.project_dir = NULL)
+ } else {
+ proj <- paste0("Project: ", basename(pdir)) %>%
+ {
+ if (nchar(.) > 35) paste0(strtrim(., 31), " ...") else .
+ }
+ options(radiant.project_dir = pdir)
+ options(radiant.launch_dir = pdir)
+ }
+
+ proj_brand <- tags$span(class = "nav navbar-brand navbar-right", proj)
+ navbar_ <- htmltools::tagQuery(navbar)$find(".navbar-collapse")$append(proj_brand)$allTags()
+ htmltools::attachDependencies(navbar_, htmltools::findDependencies(navbar))
+}
+
+if (getOption("radiant.shinyFiles", FALSE)) {
+ if (!radiant.data::is.empty(getOption("radiant.sf_volumes", "")) && radiant.data::is.empty(getOption("radiant.project_dir"))) {
+ launch_dir <- getOption("radiant.launch_dir", default = radiant.data::find_home())
+ if (!launch_dir %in% getOption("radiant.sf_volumes", "")) {
+ sf_volumes <- c(setNames(launch_dir, basename(launch_dir)), getOption("radiant.sf_volumes", ""))
+ options(radiant.sf_volumes = sf_volumes)
+ rm(sf_volumes)
+ } else if (!launch_dir == getOption("radiant.sf_volumes", "")[1]) {
+ dir_ind <- which(getOption("radiant.sf_volumes") == launch_dir)[1]
+ options(radiant.sf_volumes = c(getOption("radiant.sf_volumes")[dir_ind], getOption("radiant.sf_volumes")[-dir_ind]))
+ rm(dir_ind)
+ }
+ rm(launch_dir)
+ }
+ if (radiant.data::is.empty(getOption("radiant.launch_dir"))) {
+ if (radiant.data::is.empty(getOption("radiant.project_dir"))) {
+ options(radiant.launch_dir = radiant.data::find_home())
+ options(radiant.project_dir = getOption("radiant.launch_dir"))
+ } else {
+ options(radiant.launch_dir = getOption("radiant.project_dir"))
+ }
+ }
+
+ if (radiant.data::is.empty(getOption("radiant.project_dir"))) {
+ options(radiant.project_dir = getOption("radiant.launch_dir"))
+ } else {
+ options(radiant.launch_dir = getOption("radiant.project_dir"))
+ }
+
+ dbdir <- try(radiant.data::find_dropbox(), silent = TRUE)
+ dbdir <- if (inherits(dbdir, "try-error")) "" else paste0(dbdir, "/")
+ options(radiant.dropbox_dir = dbdir)
+ rm(dbdir)
+
+ gddir <- try(radiant.data::find_gdrive(), silent = TRUE)
+ gddir <- if (inherits(gddir, "try-error")) "" else paste0(gddir, "/")
+ options(radiant.gdrive_dir = gddir)
+ rm(gddir)
+} else {
+ options(radiant.launch_dir = radiant.data::find_home())
+ options(radiant.project_dir = getOption("radiant.launch_dir"))
+}
+
+## formatting data.frames printed in Report > Rmd and Report > R
+knit_print.data.frame <- function(x, ...) {
+ paste(c("", "", knitr::kable(x)), collapse = "\n") %>%
+ knitr::asis_output()
+}
+
+## not clear why this doesn't work
+# knit_print.data.frame = function(x, ...) {
+# res <- rmarkdown:::print.paged_df(x)
+# knitr::asis_output(res)
+# knitr::asis_output(
+# rmarkdown:::paged_table_html(x),
+# meta = list(dependencies = rmarkdown:::html_dependency_pagedtable())
+# )
+# }
+
+## not clear why this doesn't work
+## https://github.com/yihui/knitr/issues/1399
+# knit_print.datatables <- function(x, ...) {
+# res <- shiny::knit_print.shiny.render.function(
+# DT::renderDataTable(x)
+# )
+# knitr::asis_output(res)
+# }
+
+# registerS3method("knitknit_print", "datatables", knit_print.datatables)
+# knit_print.datatables <- function(x, ...) {
+# # res <- shiny::knit_print.shiny.render.function(
+# # shiny::knit_print.shiny.render.function(
+# DT::renderDataTable(x)
+# # )
+# # knitr::asis_output(res)
+# }
+
+# knit_print.datatables <- function(x, ...) {
+# # shiny::knit_print.shiny.render.function(
+# res <- shiny::knit_print.shiny.render.function(
+# DT::renderDataTable(x)
+# )
+# knitr::asis_output(res)
+# }
+
+load_html2canvas <- function() {
+ # adapted from https://github.com/yonicd/snapper/blob/master/R/load.R
+ # SRC URL "https://html2canvas.hertzen.com/dist/html2canvas.min.js"
+ asset_src <- "assets/html2canvas/"
+ asset_script <- "html2canvas.min.js"
+ dir.exists(asset_src)
+ shiny::tagList(
+ htmltools::htmlDependency(
+ name = "html2canvas-js",
+ version = "1.4.1",
+ src = asset_src,
+ script = asset_script,
+ package = "radiant.data"
+ )
+ )
+}
+
+options(
+ radiant.nav_ui =
+ list(
+ windowTitle = i18n$t("Radiant for R"),
+ id = "nav_radiant",
+ theme = getOption("radiant.theme"),
+ inverse = TRUE,
+ collapsible = TRUE,
+ position = "fixed-top",
+ tabPanel(i18n$t("Data"), withMathJax(), uiOutput("ui_data"), load_html2canvas())
+ )
+)
+
+options(
+ radiant.shared_ui =
+ tagList(
+ navbarMenu(
+ i18n$t("Report"),
+ tabPanel("Rmd",
+ uiOutput("rmd_view"),
+ uiOutput("report_rmd"),
+ icon = icon("edit", verify_fa = FALSE)
+ ),
+ tabPanel("R",
+ uiOutput("r_view"),
+ uiOutput("report_r"),
+ icon = icon("code", verify_fa = FALSE)
+ )
+ ),
+ navbarMenu("",
+ icon = icon("save", verify_fa = FALSE),
+ ## inspiration for uploading state https://stackoverflow.com/a/11406690/1974918
+ ## see also function in www/js/run_return.js
+ "Server",
+ tabPanel(actionLink("state_save_link", i18n$t("Save radiant state file"), icon = icon("download", verify_fa = FALSE))),
+ tabPanel(actionLink("state_load_link", i18n$t("Load radiant state file"), icon = icon("upload", verify_fa = FALSE))),
+ tabPanel(actionLink("state_share", i18n$t("Share radiant state"), icon = icon("share", verify_fa = FALSE))),
+ tabPanel(i18n$t("View radiant state"), uiOutput("state_view"), icon = icon("user", verify_fa = FALSE)),
+ "----", "Local",
+ tabPanel(downloadLink("state_download", tagList(icon("download", verify_fa = FALSE), i18n$t("Download radiant state file")))),
+ tabPanel(actionLink("state_upload_link", i18n$t("Upload radiant state file"), icon = icon("upload", verify_fa = FALSE)))
+ ),
+
+ ## stop app *and* close browser window
+ navbarMenu("",
+ icon = icon("power-off", verify_fa = FALSE),
+ tabPanel(
+ actionLink(
+ "stop_radiant", i18n$t("Stop"),
+ icon = icon("stop", verify_fa = FALSE),
+ onclick = "setTimeout(function(){window.close();}, 100);"
+ )
+ ),
+ tabPanel(tags$a(
+ id = "refresh_radiant", href = "#", class = "action-button",
+ list(icon("sync", verify_fa = FALSE), i18n$t("Refresh")), onclick = "window.location.reload();"
+ )),
+ ## had to remove class = "action-button" to make this work
+ tabPanel(tags$a(
+ id = "new_session", href = "./", target = "_blank",
+ list(icon("plus", verify_fa = FALSE), i18n$t("New session"))
+ ))
+ )
+ )
+)
+
+## cleanup the global environment if stop button is pressed in Rstudio
+## based on barbara's reply to
+## https://community.rstudio.com/t/rstudio-viewer-window-not-closed-on-shiny-stopapp/4158/7?u=vnijs
+onStop(function() {
+ ## don't run if the stop button was pressed in Radiant
+ if (!exists("r_data")) {
+ unlink("~/r_figures/", recursive = TRUE)
+ clean_up_list <- c(
+ "r_sessions", "help_menu", "make_url_patterns", "import_fs",
+ "init_data", "navbar_proj", "knit_print.data.frame", "withMathJax",
+ "Dropbox", "sf_volumes", "GoogleDrive", "bslib_current_version",
+ "has_bslib_theme", "load_html2canvas"
+ )
+ suppressWarnings(
+ suppressMessages({
+ res <- try(sapply(clean_up_list, function(x) if (exists(x, envir = .GlobalEnv)) rm(list = x, envir = .GlobalEnv)), silent = TRUE)
+ rm(res)
+ })
+ )
+ options(radiant.launch_dir = NULL)
+ options(radiant.project_dir = NULL)
+ message("Stopped Radiant\n")
+ stopApp()
+ }
+})
+
+## Show NA and Inf in DT tables
+## https://github.com/rstudio/DT/pull/513
+## See also https://github.com/rstudio/DT/issues/533
+## Waiting for DT.OPTION for TOJSON_ARGS
+# options(htmlwidgets.TOJSON_ARGS = list(na = "string"))
+# options("DT.TOJSON_ARGS" = list(na = "string"))
+## Sorting on client-side would be as a string, not a numeric
+## https://github.com/rstudio/DT/pull/536#issuecomment-385223433
+
+if (getRversion() < "4.4.0") `%||%` <- function(x, y) if (is.null(x)) y else x
diff --git a/radiant.data/inst/app/init.R b/radiant.data/inst/app/init.R
new file mode 100644
index 0000000000000000000000000000000000000000..207e4a4b3c03bd936fedd480c9683ef2754c958c
--- /dev/null
+++ b/radiant.data/inst/app/init.R
@@ -0,0 +1,364 @@
+################################################################################
+## functions to set initial values and take information from r_state
+## when available
+################################################################################
+
+## useful options for debugging
+# options(shiny.trace = TRUE)
+# options(shiny.error = recover)
+# options(warn = 2)
+
+if (isTRUE(getOption("radiant.shinyFiles", FALSE))) {
+ if (isTRUE(Sys.getenv("RSTUDIO") == "") && isTRUE(Sys.getenv("SHINY_PORT") != "")) {
+ ## Users not on Rstudio will only get access to pre-specified volumes
+ sf_volumes <- getOption("radiant.sf_volumes", "")
+ } else {
+ if (getOption("radiant.project_dir", "") == "") {
+ sf_volumes <- getOption("radiant.launch_dir") %>%
+ {
+ set_names(., basename(.))
+ }
+ } else {
+ sf_volumes <- getOption("radiant.project_dir") %>%
+ {
+ set_names(., basename(.))
+ }
+ }
+ home <- radiant.data::find_home()
+ if (home != sf_volumes) {
+ sf_volumes <- c(sf_volumes, home) %>% set_names(c(names(sf_volumes), "Home"))
+ } else {
+ sf_volumes <- c(Home = home)
+ }
+ if (sum(nzchar(getOption("radiant.sf_volumes", ""))) > 0) {
+ sf_volumes <- getOption("radiant.sf_volumes") %>%
+ {
+ c(sf_volumes, .[!. %in% sf_volumes])
+ }
+ }
+ missing_names <- is.na(names(sf_volumes))
+ if (sum(missing_names) > 0) {
+ sf_volumes[missing_names] <- basename(sf_volumes[missing_names])
+ }
+ }
+}
+
+remove_session_files <- function(st = Sys.time()) {
+ fl <- list.files(
+ normalizePath("~/.radiant.sessions/"),
+ pattern = "*.state.rda",
+ full.names = TRUE
+ )
+
+ for (f in fl) {
+ if (difftime(st, file.mtime(f), units = "days") > 7) {
+ unlink(f, force = TRUE)
+ }
+ }
+}
+
+remove_session_files()
+
+## from Joe Cheng's https://github.com/jcheng5/shiny-resume/blob/master/session.R
+isolate({
+ prevSSUID <- parseQueryString(session$clientData$url_search)[["SSUID"]]
+})
+
+most_recent_session_file <- function() {
+ fl <- list.files(
+ normalizePath("~/.radiant.sessions/"),
+ pattern = "*.state.rda",
+ full.names = TRUE
+ )
+
+ if (length(fl) > 0) {
+ data.frame(fn = fl, dt = file.mtime(fl), stringsAsFactors = FALSE) %>%
+ arrange(desc(dt)) %>%
+ slice(1) %>%
+ .[["fn"]] %>%
+ as.character() %>%
+ basename() %>%
+ gsub("r_(.*).state.rda", "\\1", .)
+ } else {
+ NULL
+ }
+}
+
+## set the session id
+r_ssuid <- if (getOption("radiant.local")) {
+ if (is.null(prevSSUID)) {
+ mrsf <- most_recent_session_file()
+ paste0("local-", shiny:::createUniqueId(3))
+ } else {
+ mrsf <- "0000"
+ prevSSUID
+ }
+} else {
+ ifelse(is.null(prevSSUID), shiny:::createUniqueId(5), prevSSUID)
+}
+
+## (re)start the session and push the id into the url
+session$sendCustomMessage("session_start", r_ssuid)
+
+## identify the shiny environment
+## deprecated - will be removed in future version
+r_environment <- session$token
+
+r_info_legacy <- function() {
+ r_info_elements <- c(
+ "datasetlist", "dtree_list", "pvt_rows", "nav_radiant",
+ "plot_height", "plot_width", "filter_error", "cmb_error"
+ ) %>%
+ c(paste0(r_data[["datasetlist"]], "_descr"))
+ r_info <- reactiveValues()
+ for (i in r_info_elements) {
+ r_info[[i]] <- r_data[[i]]
+ }
+ suppressWarnings(rm(list = r_info_elements, envir = r_data))
+ r_info
+}
+
+## load for previous state if available but look in global memory first
+if (isTRUE(getOption("radiant.local")) && exists("r_data", envir = .GlobalEnv)) {
+ r_data <- if (is.list(r_data)) list2env(r_data, envir = new.env()) else r_data
+ if (exists("r_info")) {
+ r_info <- do.call(reactiveValues, r_info)
+ } else {
+ r_info <- r_info_legacy()
+ }
+ r_state <- if (exists("r_state")) r_state else list()
+ suppressWarnings(rm(r_data, r_state, r_info, envir = .GlobalEnv))
+} else if (isTRUE(getOption("radiant.local")) && !is.null(r_sessions[[r_ssuid]]$r_data)) {
+ r_data <- r_sessions[[r_ssuid]]$r_data %>%
+ {
+ if (is.list(.)) list2env(., envir = new.env()) else .
+ }
+ if (is.null(r_sessions[[r_ssuid]]$r_info)) {
+ r_info <- r_info_legacy()
+ } else {
+ r_info <- do.call(reactiveValues, r_sessions[[r_ssuid]]$r_info)
+ }
+ r_state <- r_sessions[[r_ssuid]]$r_state
+} else if (file.exists(paste0("~/.radiant.sessions/r_", r_ssuid, ".state.rda"))) {
+ ## read from file if not in global
+ fn <- paste0(normalizePath("~/.radiant.sessions"), "/r_", r_ssuid, ".state.rda")
+ rs <- new.env(emptyenv())
+ try(load(fn, envir = rs), silent = TRUE)
+ if (inherits(rs, "try-error")) {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ r_state <- list()
+ } else {
+ if (length(rs$r_data) == 0) {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ } else {
+ r_data <- rs$r_data %>%
+ {
+ if (is.list(.)) list2env(., envir = new.env()) else .
+ }
+ if (is.null(rs$r_info)) {
+ r_info <- r_info_legacy()
+ } else {
+ r_info <- do.call(reactiveValues, rs$r_info)
+ }
+ }
+ if (length(rs$r_state) == 0) {
+ r_state <- list()
+ } else {
+ r_state <- rs$r_state
+ }
+ }
+
+ unlink(fn, force = TRUE)
+ rm(rs)
+} else if (isTRUE(getOption("radiant.local")) && file.exists(paste0("~/.radiant.sessions/r_", mrsf, ".state.rda"))) {
+ ## restore from local folder but assign new ssuid
+ fn <- paste0(normalizePath("~/.radiant.sessions"), "/r_", mrsf, ".state.rda")
+ rs <- new.env(emptyenv())
+ try(load(fn, envir = rs), silent = TRUE)
+ if (inherits(rs, "try-error")) {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ r_state <- list()
+ } else {
+ if (length(rs$r_data) == 0) {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ } else {
+ r_data <- rs$r_data %>%
+ {
+ if (is.list(.)) list2env(., envir = new.env()) else .
+ }
+ r_info <- if (length(rs$r_info) == 0) {
+ r_info <- r_info_legacy()
+ } else {
+ do.call(reactiveValues, rs$r_info)
+ }
+ }
+ r_state <- if (length(rs$r_state) == 0) list() else rs$r_state
+ }
+
+ ## don't navigate to same tab in case the app locks again
+ r_state$nav_radiant <- NULL
+ unlink(fn, force = TRUE)
+ rm(rs)
+} else {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ r_state <- list()
+}
+
+isolate({
+ for (ds in r_info[["datasetlist"]]) {
+ if (exists(ds, envir = r_data) && !bindingIsActive(as.symbol(ds), env = r_data)) {
+ shiny::makeReactiveBinding(ds, env = r_data)
+ }
+ }
+ for (dt in r_info[["dtree_list"]]) {
+ if (exists(dt, envir = r_data)) {
+ r_data[[dt]] <- add_class(r_data[[dt]], "dtree")
+ if (!bindingIsActive(as.symbol(dt), env = r_data)) {
+ shiny::makeReactiveBinding(dt, env = r_data)
+ }
+ }
+ }
+})
+
+## legacy, to deal with state files created before
+## Report > Rmd and Report > R name change
+if (isTRUE(r_state$nav_radiant == "Code")) {
+ r_state$nav_radiant <- "R"
+} else if (isTRUE(r_state$nav_radiant == "Report")) {
+ r_state$nav_radiant <- "Rmd"
+}
+
+## legacy, to deal with radio buttons that were in Data > Pivot
+if (!is.null(r_state$pvt_type)) {
+ if (isTRUE(r_state$pvt_type == "fill")) {
+ r_state$pvt_type <- TRUE
+ } else {
+ r_state$pvt_type <- FALSE
+ }
+}
+
+## legacy, to deal with state files created before
+## name change to rmd_edit
+if (!is.null(r_state$rmd_report) && is.null(r_state$rmd_edit)) {
+ r_state$rmd_edit <- r_state$rmd_report
+ r_state$rmd_report <- NULL
+}
+
+if (length(r_state$rmd_edit) > 0) {
+ r_state$rmd_edit <- r_state$rmd_edit %>% radiant.data::fix_smart()
+}
+
+## legacy, to deal with state files created before
+## name change to rmd_edit
+if (!is.null(r_state$rcode_edit) && is.null(r_state$r_edit)) {
+ r_state$r_edit <- r_state$rcode_edit
+ r_state$rcode_edit <- NULL
+}
+
+## parse the url and use updateTabsetPanel to navigate to the desired tab
+## currently only works with a new or refreshed session
+observeEvent(session$clientData$url_search, {
+ url_query <- parseQueryString(session$clientData$url_search)
+ if ("url" %in% names(url_query)) {
+ r_info[["url"]] <- url_query$url
+ } else if (is.empty(r_info[["url"]])) {
+ return()
+ }
+
+ ## create an observer and suspend when done
+ url_observe <- observe({
+ if (is.null(input$dataset)) {
+ return()
+ }
+ url <- getOption("radiant.url.patterns")[[r_info[["url"]]]]
+ if (is.null(url)) {
+ ## if pattern not found suspend observer
+ url_observe$suspend()
+ return()
+ }
+ ## move through the url
+ for (u in names(url)) {
+ if (is.null(input[[u]])) {
+ return()
+ }
+ if (input[[u]] != url[[u]]) {
+ updateTabsetPanel(session, u, selected = url[[u]])
+ }
+ if (names(tail(url, 1)) == u) url_observe$suspend()
+ }
+ })
+})
+
+## keeping track of the main tab we are on
+observeEvent(input$nav_radiant, {
+ if (!input$nav_radiant %in% c("Refresh", "Stop")) {
+ r_info[["nav_radiant"]] <- input$nav_radiant
+ }
+})
+
+## Jump to the page you were on
+## only goes two layers deep at this point
+if (!is.null(r_state$nav_radiant)) {
+ ## don't return-to-the-spot if that was quit or stop
+ if (r_state$nav_radiant %in% c("Refresh", "Stop")) {
+ return()
+ }
+
+ ## naming the observer so we can suspend it when done
+ nav_observe <- observe({
+ ## needed to avoid errors when no data is available yet
+ if (is.null(input$dataset)) {
+ return()
+ }
+ updateTabsetPanel(session, "nav_radiant", selected = r_state$nav_radiant)
+
+ ## check if shiny set the main tab to the desired value
+ if (is.null(input$nav_radiant)) {
+ return()
+ }
+ if (input$nav_radiant != r_state$nav_radiant) {
+ return()
+ }
+ nav_radiant_tab <- getOption("radiant.url.list")[[r_state$nav_radiant]] %>%
+ names()
+
+ if (!is.null(nav_radiant_tab) && !is.null(r_state[[nav_radiant_tab]])) {
+ updateTabsetPanel(session, nav_radiant_tab, selected = r_state[[nav_radiant_tab]])
+ }
+
+ ## once you arrive at the desired tab suspend the observer
+ nav_observe$suspend()
+ })
+}
+
+isolate({
+ if (is.null(r_info[["plot_height"]])) r_info[["plot_height"]] <- 650
+ if (is.null(r_info[["plot_width"]])) r_info[["plot_width"]] <- 650
+})
+
+if (getOption("radiant.from.package", default = TRUE)) {
+ ## launch using installed radiant.data package
+ # radiant.data::copy_all("radiant.data")
+ cat("\nGetting radiant.data from package ...\n")
+} else {
+ ## for shiny-server and development
+ for (file in list.files("../../R", pattern = "\\.(r|R)$", full.names = TRUE)) {
+ source(file, encoding = getOption("radiant.encoding", "UTF-8"), local = TRUE)
+ }
+ cat("\nGetting radiant.data from source ...\n")
+}
+
+## Getting screen width ...
+## https://github.com/rstudio/rstudio/issues/1870
+## https://community.rstudio.com/t/rstudio-resets-width-option-when-running-shiny-app-in-viewer/3661
+observeEvent(input$get_screen_width, {
+ if (getOption("width", default = 250) != 250) options(width = 250)
+})
+
+
+radiant.data::copy_from(radiant.data, register, deregister)
diff --git a/radiant.data/inst/app/radiant.R b/radiant.data/inst/app/radiant.R
new file mode 100644
index 0000000000000000000000000000000000000000..202656858a478f849a2e50d7bb1603a3c15069f6
--- /dev/null
+++ b/radiant.data/inst/app/radiant.R
@@ -0,0 +1,1034 @@
+################################################################################
+## function to save app state on refresh or crash
+################################################################################
+
+## drop NULLs in list
+toList <- function(x) reactiveValuesToList(x) %>% .[!sapply(., is.null)]
+
+## from https://gist.github.com/hadley/5434786
+env2list <- function(x) mget(ls(x), x)
+
+is_active <- function(env = r_data) {
+ sapply(ls(envir = env), function(x) bindingIsActive(as.symbol(x), env = env))
+}
+
+## remove non-active bindings
+rem_non_active <- function(env = r_data) {
+ iact <- is_active(env = r_data)
+ rm(list = names(iact)[!iact], envir = env)
+}
+
+active2list <- function(env = r_data) {
+ iact <- is_active(env = r_data) %>% (function(x) names(x)[x])
+ if (length(iact) > 0) {
+ mget(iact, env)
+ } else {
+ list()
+ }
+}
+
+## deal with https://github.com/rstudio/shiny/issues/2065
+MRB <- function(x, env = parent.frame(), init = FALSE) {
+ if (exists(x, envir = env)) {
+ ## if the object exists and has a binding, don't do anything
+ if (!bindingIsActive(as.symbol(x), env = env)) {
+ shiny::makeReactiveBinding(x, env = env)
+ }
+ } else if (init) {
+ ## initialize a binding (and value) if object doesn't exist yet
+ shiny::makeReactiveBinding(x, env = env)
+ }
+}
+
+saveSession <- function(session = session, timestamp = FALSE, path = "~/.radiant.sessions") {
+ if (!exists("r_sessions")) {
+ return()
+ }
+ if (!dir.exists(path)) dir.create(path)
+ isolate({
+ LiveInputs <- toList(input)
+ r_state[names(LiveInputs)] <- LiveInputs
+
+ ## removing the non-active bindings
+ rem_non_active()
+
+ r_data <- env2list(r_data)
+ r_info <- toList(r_info)
+
+ r_sessions[[r_ssuid]] <- list(
+ r_data = r_data,
+ r_info = r_info,
+ r_state = r_state,
+ timestamp = Sys.time()
+ )
+
+ ## saving session information to state file
+ if (timestamp) {
+ fn <- paste0(normalizePath(path), "/r_", r_ssuid, "-", gsub("( |:)", "-", Sys.time()), ".state.rda")
+ } else {
+ fn <- paste0(normalizePath(path), "/r_", r_ssuid, ".state.rda")
+ }
+ save(r_data, r_info, r_state, file = fn)
+ })
+}
+
+observeEvent(input$refresh_radiant, {
+ if (isTRUE(getOption("radiant.local"))) {
+ fn <- normalizePath("~/.radiant.sessions")
+ file.remove(list.files(fn, full.names = TRUE))
+ } else {
+ fn <- paste0(normalizePath("~/.radiant.sessions"), "/r_", r_ssuid, ".state.rda")
+ if (file.exists(fn)) unlink(fn, force = TRUE)
+ }
+
+ try(r_ssuid <- NULL, silent = TRUE)
+})
+
+saveStateOnRefresh <- function(session = session) {
+ session$onSessionEnded(function() {
+ isolate({
+ url_query <- parseQueryString(session$clientData$url_search)
+ if (not_pressed(input$refresh_radiant) &&
+ not_pressed(input$stop_radiant) &&
+ not_pressed(input$state_load) &&
+ not_pressed(input$state_upload) &&
+ !"fixed" %in% names(url_query)) {
+ saveSession(session)
+ } else {
+ if (not_pressed(input$state_load) && not_pressed(input$state_upload)) {
+ if (exists("r_sessions")) {
+ sshhr(try(r_sessions[[r_ssuid]] <- NULL, silent = TRUE))
+ sshhr(try(rm(r_ssuid), silent = TRUE))
+ }
+ }
+ }
+ })
+ })
+}
+
+################################################################
+## functions used across tools in radiant
+################################################################
+
+## get active dataset and apply data-filter if available
+.get_data <- reactive({
+ req(input$dataset)
+
+ filter_cmd <- input$data_filter %>%
+ gsub("\\n", "", .) %>%
+ gsub("\"", "\'", .) %>%
+ fix_smart()
+
+ arrange_cmd <- input$data_arrange
+ if (!is.empty(arrange_cmd)) {
+ arrange_cmd <- arrange_cmd %>%
+ strsplit(., split = "(&|,|\\s+)") %>%
+ unlist() %>%
+ .[!. == ""] %>%
+ paste0(collapse = ", ") %>%
+ (function(x) glue("arrange(x, {x})"))
+ }
+
+ slice_cmd <- input$data_rows
+
+ if ((is.empty(filter_cmd) && is.empty(arrange_cmd) && is.empty(slice_cmd)) || input$show_filter == FALSE) {
+ isolate(r_info[["filter_error"]] <- "")
+ } else if (grepl("([^=!<>])=([^=])", filter_cmd)) {
+ isolate(r_info[["filter_error"]] <- "Invalid filter: Never use = in a filter! Use == instead (e.g., city == 'San Diego'). Update or remove the expression")
+ } else {
+ ## %>% needed here so . will be available
+ seldat <- try(
+ r_data[[input$dataset]] %>%
+ (function(x) if (!is.empty(filter_cmd)) x %>% filter(!!rlang::parse_expr(filter_cmd)) else x) %>%
+ (function(x) if (!is.empty(arrange_cmd)) eval(parse(text = arrange_cmd)) else x) %>%
+ (function(x) if (!is.empty(slice_cmd)) x %>% slice(!!rlang::parse_expr(slice_cmd)) else x),
+ silent = TRUE
+ )
+ if (inherits(seldat, "try-error")) {
+ isolate(r_info[["filter_error"]] <- paste0("Invalid input: \"", attr(seldat, "condition")$message, "\". Update or remove the expression(x)"))
+ } else {
+ isolate(r_info[["filter_error"]] <- "")
+ if ("grouped_df" %in% class(seldat)) {
+ return(droplevels(ungroup(seldat)))
+ } else {
+ return(droplevels(seldat))
+ }
+ }
+ }
+ if ("grouped_df" %in% class(r_data[[input$dataset]])) {
+ ungroup(r_data[[input$dataset]])
+ } else {
+ r_data[[input$dataset]]
+ }
+})
+
+## using a regular function to avoid a full data copy
+.get_data_transform <- function(dataset = input$dataset) {
+ if (is.null(dataset)) {
+ return()
+ }
+ if ("grouped_df" %in% class(r_data[[dataset]])) {
+ ungroup(r_data[[dataset]])
+ } else {
+ r_data[[dataset]]
+ }
+}
+
+.get_class <- reactive({
+ get_class(.get_data())
+})
+
+groupable_vars <- reactive({
+ .get_data() %>%
+ summarise_all(
+ list(
+ ~ is.factor(.) || is.logical(.) || lubridate::is.Date(.) ||
+ is.integer(.) || is.character(.) ||
+ ((length(unique(.)) / n()) < 0.30)
+ )
+ ) %>%
+ (function(x) which(x == TRUE)) %>%
+ varnames()[.]
+})
+
+groupable_vars_nonum <- reactive({
+ .get_data() %>%
+ summarise_all(
+ list(
+ ~ is.factor(.) || is.logical(.) ||
+ lubridate::is.Date(.) || is.integer(.) ||
+ is.character(.)
+ )
+ ) %>%
+ (function(x) which(x == TRUE)) %>%
+ varnames()[.]
+})
+
+## used in compare proportions, logistic, etc.
+two_level_vars <- reactive({
+ two_levs <- function(x) {
+ if (is.factor(x)) {
+ length(levels(x))
+ } else {
+ length(unique(na.omit(x)))
+ }
+ }
+ .get_data() %>%
+ summarise_all(two_levs) %>%
+ (function(x) x == 2) %>%
+ which(.) %>%
+ varnames()[.]
+})
+
+## used in visualize - don't plot Y-variables that don't vary
+varying_vars <- reactive({
+ .get_data() %>%
+ summarise_all(does_vary) %>%
+ as.logical() %>%
+ which() %>%
+ varnames()[.]
+})
+
+## getting variable names in active dataset and their class
+varnames <- reactive({
+ var_class <- .get_class()
+ req(var_class)
+ names(var_class) %>%
+ set_names(., paste0(., " {", var_class, "}"))
+})
+
+## cleaning up the arguments for data_filter and defaults passed to report
+clean_args <- function(rep_args, rep_default = list()) {
+ if (!is.null(rep_args$data_filter)) {
+ if (rep_args$data_filter == "") {
+ rep_args$data_filter <- NULL
+ } else {
+ rep_args$data_filter %<>% gsub("\\n", "", .) %>% gsub("\"", "\'", .)
+ }
+ }
+ if (is.empty(rep_args$rows)) {
+ rep_args$rows <- NULL
+ }
+ if (is.empty(rep_args$arr)) {
+ rep_args$arr <- NULL
+ }
+
+ if (length(rep_default) == 0) rep_default[names(rep_args)] <- ""
+
+ ## removing default arguments before sending to report feature
+ for (i in names(rep_args)) {
+ if (!is.language(rep_args[[i]]) && !is.call(rep_args[[i]]) && all(is.na(rep_args[[i]]))) {
+ rep_args[[i]] <- NULL
+ next
+ }
+ if (!is.symbol(rep_default[[i]]) && !is.call(rep_default[[i]]) && all(is_not(rep_default[[i]]))) next
+ if (length(rep_args[[i]]) == length(rep_default[[i]]) && !is.name(rep_default[[i]]) && all(rep_args[[i]] == rep_default[[i]])) {
+ rep_args[[i]] <- NULL
+ }
+ }
+
+ rep_args
+}
+
+## check if a variable is null or not in the selected data.frame
+not_available <- function(x) any(is.null(x)) || (sum(x %in% varnames()) < length(x))
+
+## check if a variable is null or not in the selected data.frame
+available <- function(x) !not_available(x)
+
+## check if a button was pressed
+pressed <- function(x) !is.null(x) && (is.list(x) || x > 0)
+
+## check if a button was NOT pressed
+not_pressed <- function(x) !pressed(x)
+
+## check for duplicate entries
+has_duplicates <- function(x) length(unique(x)) < length(x)
+
+## is x some type of date variable
+is_date <- function(x) inherits(x, c("Date", "POSIXlt", "POSIXct"))
+
+## drop elements from .._args variables obtained using formals
+r_drop <- function(x, drop = c("dataset", "data_filter", "arr", "rows", "envir")) x[!x %in% drop]
+
+## show a few rows of a dataframe
+show_data_snippet <- function(dataset = input$dataset, nshow = 7, title = "", filt = "", arr = "", rows = "") {
+ if (is.character(dataset) && length(dataset) == 1) dataset <- get_data(dataset, filt = filt, arr = arr, rows = rows, na.rm = FALSE, envir = r_data)
+ nr <- nrow(dataset)
+ ## avoid slice with variables outside of the df in case a column with the same
+ ## name exists
+ dataset[1:min(nshow, nr), , drop = FALSE] %>%
+ mutate_if(is_date, as.character) %>%
+ mutate_if(is.character, list(~ strtrim(., 40))) %>%
+ xtable::xtable(.) %>%
+ print(
+ type = "html", print.results = FALSE, include.rownames = FALSE,
+ sanitize.text.function = identity,
+ html.table.attributes = "class='table table-condensed table-hover snippet'"
+ ) %>%
+ paste0(title, .) %>%
+ (function(x) if (nr <= nshow) x else paste0(x, "\n")) %>%
+ enc2utf8()
+}
+
+suggest_data <- function(text = "", df_name = "diamonds") {
+ paste0(text, "要获取示例数据集,请转到数据 > 管理,从加载数据类型'下拉菜单中选择'示例',\n然后点击'加载'按钮,选择 '", df_name, "' 数据集。")
+}
+
+## function written by @wch https://github.com/rstudio/shiny/issues/781#issuecomment-87135411
+capture_plot <- function(expr, env = parent.frame()) {
+ structure(
+ list(expr = substitute(expr), env = env),
+ class = "capture_plot"
+ )
+}
+
+## function written by @wch https://github.com/rstudio/shiny/issues/781#issuecomment-87135411
+print.capture_plot <- function(x, ...) {
+ eval(x$expr, x$env)
+}
+
+################################################################
+## functions used to create Shiny in and outputs
+################################################################
+
+## textarea where the return key submits the content
+returnTextAreaInput <- function(inputId, label = NULL, rows = 2,
+ placeholder = NULL, resize = "vertical",
+ value = "") {
+ ## avoid all sorts of 'helpful' behavior from your browser
+ ## see https://stackoverflow.com/a/35514029/1974918
+ tagList(
+ tags$div(
+ # using containing element based on
+ # https://github.com/niklasvh/html2canvas/issues/2008#issuecomment-1445503369
+ tags$label(label, `for` = inputId), br(),
+ tags$textarea(
+ value,
+ id = inputId,
+ type = "text",
+ rows = rows,
+ placeholder = placeholder,
+ resize = resize,
+ autocomplete = "off",
+ autocorrect = "off",
+ autocapitalize = "off",
+ spellcheck = "false",
+ class = "returnTextArea form-control"
+ )
+ )
+ )
+}
+
+## from https://github.com/rstudio/shiny/blob/master/R/utils.R
+`%AND%` <- function(x, y) {
+ if (!all(is.null(x)) && !all(is.na(x))) {
+ if (!all(is.null(y)) && !all(is.na(y))) {
+ return(y)
+ }
+ }
+ return(NULL)
+}
+
+## using a custom version of textInput to avoid browser "smartness"
+textInput <- function(inputId, label, value = "", width = NULL,
+ placeholder = NULL, autocomplete = "off",
+ autocorrect = "off", autocapitalize = "off",
+ spellcheck = "false", ...) {
+ value <- restoreInput(id = inputId, default = value)
+ div(
+ class = "form-group shiny-input-container",
+ style = if (!is.null(width)) paste0("width: ", validateCssUnit(width), ";"),
+ label %AND% tags$label(label, `for` = inputId),
+ tags$input(
+ id = inputId,
+ type = "text",
+ class = "form-control",
+ value = value,
+ placeholder = placeholder,
+ autocomplete = autocomplete,
+ autocorrect = autocorrect,
+ autocapitalize = autocapitalize,
+ spellcheck = spellcheck,
+ ...
+ )
+ )
+}
+
+## using a custom version of textAreaInput to avoid browser "smartness"
+textAreaInput <- function(inputId, label, value = "", width = NULL,
+ height = NULL, cols = NULL, rows = NULL,
+ placeholder = NULL, resize = NULL,
+ autocomplete = "off", autocorrect = "off",
+ autocapitalize = "off", spellcheck = "true",
+ ...) {
+ value <- restoreInput(id = inputId, default = value)
+ if (!is.null(resize)) {
+ resize <- match.arg(
+ resize,
+ c("both", "none", "vertical", "horizontal")
+ )
+ }
+ style <- paste(if (!is.null(width)) {
+ paste0("width: ", validateCssUnit(width), ";")
+ }, if (!is.null(height)) {
+ paste0("height: ", validateCssUnit(height), ";")
+ }, if (!is.null(resize)) {
+ paste0("resize: ", resize, ";")
+ })
+ if (length(style) == 0) {
+ style <- NULL
+ }
+ div(
+ class = "form-group shiny-input-container",
+ label %AND% tags$label(label, `for` = inputId),
+ tags$textarea(
+ id = inputId,
+ class = "form-control",
+ placeholder = placeholder,
+ style = style,
+ rows = rows,
+ cols = cols,
+ autocomplete = autocomplete,
+ autocorrect = autocorrect,
+ autocapitalize = autocapitalize,
+ spellcheck = spellcheck,
+ ...,
+ value
+ )
+ )
+}
+
+## avoid all sorts of 'helpful' behavior from your browser
+## based on https://stackoverflow.com/a/35514029/1974918
+returnTextInput <- function(inputId, label = NULL,
+ placeholder = NULL, value = "") {
+ tagList(
+ tags$label(label, `for` = inputId),
+ tags$input(
+ id = inputId,
+ type = "text",
+ value = value,
+ placeholder = placeholder,
+ autocomplete = "off",
+ autocorrect = "off",
+ autocapitalize = "off",
+ spellcheck = "false",
+ class = "returnTextInput form-control"
+ )
+ )
+}
+
+if (getOption("radiant.shinyFiles", FALSE)) {
+ download_link <- function(id) {
+ uiOutput(paste0("ui_", id))
+ }
+
+ download_button <- function(id, ...) {
+ uiOutput(paste0("ui_", id))
+ }
+
+ download_handler <- function(id, label = "", fun = id, fn, type = "csv", caption = "Save to csv",
+ class = "", ic = "download", btn = "link", onclick = "function() none;", ...) {
+ ## create observer
+ shinyFiles::shinyFileSave(input, id, roots = sf_volumes, session = session)
+
+ ## create renderUI
+ if (btn == "link") {
+ output[[paste0("ui_", id)]] <- renderUI({
+ if (is.function(fn)) fn <- fn()
+ if (is.function(type)) type <- type()
+ shinyFiles::shinySaveLink(
+ id, label, caption,
+ filename = fn, filetype = type,
+ class = "alignright", icon = icon(ic, verify_fa = FALSE), onclick = onclick
+ )
+ })
+ } else {
+ output[[paste0("ui_", id)]] <- renderUI({
+ if (is.function(fn)) fn <- fn()
+ if (is.function(type)) type <- type()
+ shinyFiles::shinySaveButton(
+ id, label, caption,
+ filename = fn, filetype = type,
+ class = class, icon = icon("download", verify_fa = FALSE), onclick = onclick
+ )
+ })
+ }
+
+ observeEvent(input[[id]], {
+ if (is.integer(input[[id]])) {
+ return()
+ }
+ path <- shinyFiles::parseSavePath(sf_volumes, input[[id]])
+ if (!inherits(path, "try-error") && !is.empty(path$datapath)) {
+ fun(path$datapath, ...)
+ }
+ })
+ }
+} else {
+ download_link <- function(id, ...) {
+ downloadLink(id, "", class = "fa fa-download alignright", ...)
+ }
+ download_button <- function(id, label = "Save", ic = "download", class = "", ...) {
+ downloadButton(id, label, class = class, ...)
+ }
+ download_handler <- function(id, label = "", fun = id, fn, type = "csv", caption = "Save to csv",
+ class = "", ic = "download", btn = "link", ...) {
+ output[[id]] <- downloadHandler(
+ filename = function() {
+ if (is.function(fn)) fn <- fn()
+ if (is.function(type)) type <- type()
+ paste0(fn, ".", type)
+ },
+ content = function(path) {
+ fun(path, ...)
+ }
+ )
+ }
+}
+
+plot_width <- function() {
+ if (is.null(input$viz_plot_width)) r_info[["plot_width"]] else input$viz_plot_width
+}
+
+plot_height <- function() {
+ if (is.null(input$viz_plot_height)) r_info[["plot_height"]] else input$viz_plot_height
+}
+
+download_handler_plot <- function(path, plot, width = plot_width, height = plot_height) {
+ plot <- try(plot(), silent = TRUE)
+ if (inherits(plot, "try-error") || is.character(plot) || is.null(plot)) {
+ plot <- ggplot() +
+ labs(title = "Plot not available")
+ inp <- c(500, 100, 96)
+ } else {
+ inp <- 5 * c(width(), height(), 96)
+ }
+
+ png(file = path, width = inp[1], height = inp[2], res = inp[3])
+ print(plot)
+ dev.off()
+}
+
+## fun_name is a string of the main function name
+## rfun_name is a string of the reactive wrapper that calls the main function
+## out_name is the name of the output, set to fun_name by default
+register_print_output <- function(fun_name, rfun_name, out_name = fun_name) {
+ ## Generate output for the summary tab
+ output[[out_name]] <- renderPrint({
+ ## when no analysis was conducted (e.g., no variables selected)
+ fun <- get(rfun_name)()
+ if (is.character(fun)) {
+ cat(fun, "\n")
+ } else {
+ rm(fun)
+ }
+ })
+ return(invisible())
+}
+
+## fun_name is a string of the main function name
+## rfun_name is a string of the reactive wrapper that calls the main function
+## out_name is the name of the output, set to fun_name by default
+register_plot_output <- function(fun_name, rfun_name, out_name = fun_name,
+ width_fun = "plot_width", height_fun = "plot_height") {
+ ## Generate output for the plots tab
+ output[[out_name]] <- renderPlot(
+ {
+ ## when no analysis was conducted (e.g., no variables selected)
+ p <- get(rfun_name)()
+ if (is_not(p) || is.empty(p)) p <- "Nothing to plot ...\nSelect plots to show or re-run the calculations"
+ if (is.character(p)) {
+ plot(
+ x = 1, type = "n", main = paste0("\n\n\n\n\n\n\n\n", p),
+ axes = FALSE, xlab = "", ylab = "", cex.main = .9
+ )
+ } else {
+ print(p)
+ }
+ },
+ width = get(width_fun),
+ height = get(height_fun),
+ res = 96
+ )
+
+ return(invisible())
+}
+
+stat_tab_panel <- function(menu, tool, tool_ui, output_panels,
+ data = input$dataset) {
+ sidebarLayout(
+ sidebarPanel(
+ wellPanel(
+ HTML(paste("
")
+
+#####################################
+## url processing to share results
+#####################################
+
+## relevant links
+# http://stackoverflow.com/questions/25306519/shiny-saving-url-state-subpages-and-tabs/25385474#25385474
+# https://groups.google.com/forum/#!topic/shiny-discuss/Xgxq08N8HBE
+# https://gist.github.com/jcheng5/5427d6f264408abf3049
+
+## try http://127.0.0.1:3174/?url=multivariate/conjoint/plot/&SSUID=local
+options(
+ radiant.url.list =
+ list("Data" = list("tabs_data" = list(
+ "Manage" = "data/",
+ "View" = "data/view/",
+ "Visualize" = "data/visualize/",
+ "Pivot" = "data/pivot/",
+ "Explore" = "data/explore/",
+ "Transform" = "data/transform/",
+ "Combine" = "data/combine/",
+ "Rmd" = "report/rmd/",
+ "R" = "report/r/"
+ )))
+)
+
+make_url_patterns <- function(url_list = getOption("radiant.url.list"),
+ url_patterns = list()) {
+ for (i in names(url_list)) {
+ res <- url_list[[i]]
+ if (!is.list(res)) {
+ url_patterns[[res]] <- list("nav_radiant" = i)
+ } else {
+ tabs <- names(res)
+ for (j in names(res[[tabs]])) {
+ url <- res[[tabs]][[j]]
+ url_patterns[[url]] <- setNames(list(i, j), c("nav_radiant", tabs))
+ }
+ }
+ }
+ url_patterns
+}
+
+## generate url patterns
+options(radiant.url.patterns = make_url_patterns())
+
+## installed packages versions
+tmp <- grep("radiant\\.", installed.packages()[, "Package"], value = TRUE)
+if ("radiant" %in% installed.packages()) {
+ tmp <- c("radiant" = "radiant", tmp)
+}
+
+if (length(tmp) > 0) {
+ radiant.versions <- sapply(names(tmp), function(x) paste(x, paste(packageVersion(x), sep = "."))) %>% unique()
+ if ("shiny" %in% installed.packages()) {
+ radiant.versions <- c(radiant.versions, paste("shiny ", packageVersion("shiny")))
+ }
+} else {
+ radiant.versions <- "Unknown"
+}
+
+options(radiant.versions = paste(radiant.versions, collapse = ", "))
+rm(tmp, radiant.versions)
+
+if (is.null(getOption("radiant.theme", default = NULL))) {
+ options(radiant.theme = bslib::bs_theme(version = 4))
+}
+
+## bslib theme and version
+has_bslib_theme <- function() {
+ if (rlang::is_installed("bslib")) bslib::is_bs_theme(getOption("radiant.theme")) else FALSE
+}
+
+bslib_current_version <- function() {
+ if (rlang::is_installed("bslib")) bslib::theme_version(getOption("radiant.theme", default = bslib::bs_theme(version = 4)))
+}
+
+navbar_proj <- function(navbar) {
+ pdir <- radiant.data::find_project(mess = FALSE)
+ if (radiant.data::is.empty(pdir)) {
+ if (getOption("radiant.shinyFiles", FALSE) && !radiant.data::is.empty(getOption("radiant.sf_volumes", ""))) {
+ proj <- paste0(i18n$t("Base dir: "), names(getOption("radiant.sf_volumes"))[1])
+ } else {
+ proj <- "Project: (None)"
+ }
+ options(radiant.project_dir = NULL)
+ } else {
+ proj <- paste0("Project: ", basename(pdir)) %>%
+ {
+ if (nchar(.) > 35) paste0(strtrim(., 31), " ...") else .
+ }
+ options(radiant.project_dir = pdir)
+ options(radiant.launch_dir = pdir)
+ }
+
+ proj_brand <- tags$span(class = "nav navbar-brand navbar-right", proj)
+ navbar_ <- htmltools::tagQuery(navbar)$find(".navbar-collapse")$append(proj_brand)$allTags()
+ htmltools::attachDependencies(navbar_, htmltools::findDependencies(navbar))
+}
+
+if (getOption("radiant.shinyFiles", FALSE)) {
+ if (!radiant.data::is.empty(getOption("radiant.sf_volumes", "")) && radiant.data::is.empty(getOption("radiant.project_dir"))) {
+ launch_dir <- getOption("radiant.launch_dir", default = radiant.data::find_home())
+ if (!launch_dir %in% getOption("radiant.sf_volumes", "")) {
+ sf_volumes <- c(setNames(launch_dir, basename(launch_dir)), getOption("radiant.sf_volumes", ""))
+ options(radiant.sf_volumes = sf_volumes)
+ rm(sf_volumes)
+ } else if (!launch_dir == getOption("radiant.sf_volumes", "")[1]) {
+ dir_ind <- which(getOption("radiant.sf_volumes") == launch_dir)[1]
+ options(radiant.sf_volumes = c(getOption("radiant.sf_volumes")[dir_ind], getOption("radiant.sf_volumes")[-dir_ind]))
+ rm(dir_ind)
+ }
+ rm(launch_dir)
+ }
+ if (radiant.data::is.empty(getOption("radiant.launch_dir"))) {
+ if (radiant.data::is.empty(getOption("radiant.project_dir"))) {
+ options(radiant.launch_dir = radiant.data::find_home())
+ options(radiant.project_dir = getOption("radiant.launch_dir"))
+ } else {
+ options(radiant.launch_dir = getOption("radiant.project_dir"))
+ }
+ }
+
+ if (radiant.data::is.empty(getOption("radiant.project_dir"))) {
+ options(radiant.project_dir = getOption("radiant.launch_dir"))
+ } else {
+ options(radiant.launch_dir = getOption("radiant.project_dir"))
+ }
+
+ dbdir <- try(radiant.data::find_dropbox(), silent = TRUE)
+ dbdir <- if (inherits(dbdir, "try-error")) "" else paste0(dbdir, "/")
+ options(radiant.dropbox_dir = dbdir)
+ rm(dbdir)
+
+ gddir <- try(radiant.data::find_gdrive(), silent = TRUE)
+ gddir <- if (inherits(gddir, "try-error")) "" else paste0(gddir, "/")
+ options(radiant.gdrive_dir = gddir)
+ rm(gddir)
+} else {
+ options(radiant.launch_dir = radiant.data::find_home())
+ options(radiant.project_dir = getOption("radiant.launch_dir"))
+}
+
+## formatting data.frames printed in Report > Rmd and Report > R
+knit_print.data.frame <- function(x, ...) {
+ paste(c("", "", knitr::kable(x)), collapse = "\n") %>%
+ knitr::asis_output()
+}
+
+## not clear why this doesn't work
+# knit_print.data.frame = function(x, ...) {
+# res <- rmarkdown:::print.paged_df(x)
+# knitr::asis_output(res)
+# knitr::asis_output(
+# rmarkdown:::paged_table_html(x),
+# meta = list(dependencies = rmarkdown:::html_dependency_pagedtable())
+# )
+# }
+
+## not clear why this doesn't work
+## https://github.com/yihui/knitr/issues/1399
+# knit_print.datatables <- function(x, ...) {
+# res <- shiny::knit_print.shiny.render.function(
+# DT::renderDataTable(x)
+# )
+# knitr::asis_output(res)
+# }
+
+# registerS3method("knitknit_print", "datatables", knit_print.datatables)
+# knit_print.datatables <- function(x, ...) {
+# # res <- shiny::knit_print.shiny.render.function(
+# # shiny::knit_print.shiny.render.function(
+# DT::renderDataTable(x)
+# # )
+# # knitr::asis_output(res)
+# }
+
+# knit_print.datatables <- function(x, ...) {
+# # shiny::knit_print.shiny.render.function(
+# res <- shiny::knit_print.shiny.render.function(
+# DT::renderDataTable(x)
+# )
+# knitr::asis_output(res)
+# }
+
+load_html2canvas <- function() {
+ # adapted from https://github.com/yonicd/snapper/blob/master/R/load.R
+ # SRC URL "https://html2canvas.hertzen.com/dist/html2canvas.min.js"
+ asset_src <- "assets/html2canvas/"
+ asset_script <- "html2canvas.min.js"
+ dir.exists(asset_src)
+ shiny::tagList(
+ htmltools::htmlDependency(
+ name = "html2canvas-js",
+ version = "1.4.1",
+ src = asset_src,
+ script = asset_script,
+ package = "radiant.data"
+ )
+ )
+}
+
+options(
+ radiant.nav_ui =
+ list(
+ windowTitle = i18n$t("Radiant for R"),
+ id = "nav_radiant",
+ theme = getOption("radiant.theme"),
+ inverse = TRUE,
+ collapsible = TRUE,
+ position = "fixed-top",
+ tabPanel(i18n$t("Data"), withMathJax(), uiOutput("ui_data"), load_html2canvas())
+ )
+)
+
+options(
+ radiant.shared_ui =
+ tagList(
+ navbarMenu(
+ i18n$t("Report"),
+ tabPanel("Rmd",
+ uiOutput("rmd_view"),
+ uiOutput("report_rmd"),
+ icon = icon("edit", verify_fa = FALSE)
+ ),
+ tabPanel("R",
+ uiOutput("r_view"),
+ uiOutput("report_r"),
+ icon = icon("code", verify_fa = FALSE)
+ )
+ ),
+ navbarMenu("",
+ icon = icon("save", verify_fa = FALSE),
+ ## inspiration for uploading state https://stackoverflow.com/a/11406690/1974918
+ ## see also function in www/js/run_return.js
+ "Server",
+ tabPanel(actionLink("state_save_link", i18n$t("Save radiant state file"), icon = icon("download", verify_fa = FALSE))),
+ tabPanel(actionLink("state_load_link", i18n$t("Load radiant state file"), icon = icon("upload", verify_fa = FALSE))),
+ tabPanel(actionLink("state_share", i18n$t("Share radiant state"), icon = icon("share", verify_fa = FALSE))),
+ tabPanel(i18n$t("View radiant state"), uiOutput("state_view"), icon = icon("user", verify_fa = FALSE)),
+ "----", "Local",
+ tabPanel(downloadLink("state_download", tagList(icon("download", verify_fa = FALSE), i18n$t("Download radiant state file")))),
+ tabPanel(actionLink("state_upload_link", i18n$t("Upload radiant state file"), icon = icon("upload", verify_fa = FALSE)))
+ ),
+
+ ## stop app *and* close browser window
+ navbarMenu("",
+ icon = icon("power-off", verify_fa = FALSE),
+ tabPanel(
+ actionLink(
+ "stop_radiant", i18n$t("Stop"),
+ icon = icon("stop", verify_fa = FALSE),
+ onclick = "setTimeout(function(){window.close();}, 100);"
+ )
+ ),
+ tabPanel(tags$a(
+ id = "refresh_radiant", href = "#", class = "action-button",
+ list(icon("sync", verify_fa = FALSE), i18n$t("Refresh")), onclick = "window.location.reload();"
+ )),
+ ## had to remove class = "action-button" to make this work
+ tabPanel(tags$a(
+ id = "new_session", href = "./", target = "_blank",
+ list(icon("plus", verify_fa = FALSE), i18n$t("New session"))
+ ))
+ )
+ )
+)
+
+## cleanup the global environment if stop button is pressed in Rstudio
+## based on barbara's reply to
+## https://community.rstudio.com/t/rstudio-viewer-window-not-closed-on-shiny-stopapp/4158/7?u=vnijs
+onStop(function() {
+ ## don't run if the stop button was pressed in Radiant
+ if (!exists("r_data")) {
+ unlink("~/r_figures/", recursive = TRUE)
+ clean_up_list <- c(
+ "r_sessions", "help_menu", "make_url_patterns", "import_fs",
+ "init_data", "navbar_proj", "knit_print.data.frame", "withMathJax",
+ "Dropbox", "sf_volumes", "GoogleDrive", "bslib_current_version",
+ "has_bslib_theme", "load_html2canvas"
+ )
+ suppressWarnings(
+ suppressMessages({
+ res <- try(sapply(clean_up_list, function(x) if (exists(x, envir = .GlobalEnv)) rm(list = x, envir = .GlobalEnv)), silent = TRUE)
+ rm(res)
+ })
+ )
+ options(radiant.launch_dir = NULL)
+ options(radiant.project_dir = NULL)
+ message("Stopped Radiant\n")
+ stopApp()
+ }
+})
+
+## Show NA and Inf in DT tables
+## https://github.com/rstudio/DT/pull/513
+## See also https://github.com/rstudio/DT/issues/533
+## Waiting for DT.OPTION for TOJSON_ARGS
+# options(htmlwidgets.TOJSON_ARGS = list(na = "string"))
+# options("DT.TOJSON_ARGS" = list(na = "string"))
+## Sorting on client-side would be as a string, not a numeric
+## https://github.com/rstudio/DT/pull/536#issuecomment-385223433
+
+if (getRversion() < "4.4.0") `%||%` <- function(x, y) if (is.null(x)) y else x
diff --git a/radiant.data/inst/app/init.R b/radiant.data/inst/app/init.R
new file mode 100644
index 0000000000000000000000000000000000000000..207e4a4b3c03bd936fedd480c9683ef2754c958c
--- /dev/null
+++ b/radiant.data/inst/app/init.R
@@ -0,0 +1,364 @@
+################################################################################
+## functions to set initial values and take information from r_state
+## when available
+################################################################################
+
+## useful options for debugging
+# options(shiny.trace = TRUE)
+# options(shiny.error = recover)
+# options(warn = 2)
+
+if (isTRUE(getOption("radiant.shinyFiles", FALSE))) {
+ if (isTRUE(Sys.getenv("RSTUDIO") == "") && isTRUE(Sys.getenv("SHINY_PORT") != "")) {
+ ## Users not on Rstudio will only get access to pre-specified volumes
+ sf_volumes <- getOption("radiant.sf_volumes", "")
+ } else {
+ if (getOption("radiant.project_dir", "") == "") {
+ sf_volumes <- getOption("radiant.launch_dir") %>%
+ {
+ set_names(., basename(.))
+ }
+ } else {
+ sf_volumes <- getOption("radiant.project_dir") %>%
+ {
+ set_names(., basename(.))
+ }
+ }
+ home <- radiant.data::find_home()
+ if (home != sf_volumes) {
+ sf_volumes <- c(sf_volumes, home) %>% set_names(c(names(sf_volumes), "Home"))
+ } else {
+ sf_volumes <- c(Home = home)
+ }
+ if (sum(nzchar(getOption("radiant.sf_volumes", ""))) > 0) {
+ sf_volumes <- getOption("radiant.sf_volumes") %>%
+ {
+ c(sf_volumes, .[!. %in% sf_volumes])
+ }
+ }
+ missing_names <- is.na(names(sf_volumes))
+ if (sum(missing_names) > 0) {
+ sf_volumes[missing_names] <- basename(sf_volumes[missing_names])
+ }
+ }
+}
+
+remove_session_files <- function(st = Sys.time()) {
+ fl <- list.files(
+ normalizePath("~/.radiant.sessions/"),
+ pattern = "*.state.rda",
+ full.names = TRUE
+ )
+
+ for (f in fl) {
+ if (difftime(st, file.mtime(f), units = "days") > 7) {
+ unlink(f, force = TRUE)
+ }
+ }
+}
+
+remove_session_files()
+
+## from Joe Cheng's https://github.com/jcheng5/shiny-resume/blob/master/session.R
+isolate({
+ prevSSUID <- parseQueryString(session$clientData$url_search)[["SSUID"]]
+})
+
+most_recent_session_file <- function() {
+ fl <- list.files(
+ normalizePath("~/.radiant.sessions/"),
+ pattern = "*.state.rda",
+ full.names = TRUE
+ )
+
+ if (length(fl) > 0) {
+ data.frame(fn = fl, dt = file.mtime(fl), stringsAsFactors = FALSE) %>%
+ arrange(desc(dt)) %>%
+ slice(1) %>%
+ .[["fn"]] %>%
+ as.character() %>%
+ basename() %>%
+ gsub("r_(.*).state.rda", "\\1", .)
+ } else {
+ NULL
+ }
+}
+
+## set the session id
+r_ssuid <- if (getOption("radiant.local")) {
+ if (is.null(prevSSUID)) {
+ mrsf <- most_recent_session_file()
+ paste0("local-", shiny:::createUniqueId(3))
+ } else {
+ mrsf <- "0000"
+ prevSSUID
+ }
+} else {
+ ifelse(is.null(prevSSUID), shiny:::createUniqueId(5), prevSSUID)
+}
+
+## (re)start the session and push the id into the url
+session$sendCustomMessage("session_start", r_ssuid)
+
+## identify the shiny environment
+## deprecated - will be removed in future version
+r_environment <- session$token
+
+r_info_legacy <- function() {
+ r_info_elements <- c(
+ "datasetlist", "dtree_list", "pvt_rows", "nav_radiant",
+ "plot_height", "plot_width", "filter_error", "cmb_error"
+ ) %>%
+ c(paste0(r_data[["datasetlist"]], "_descr"))
+ r_info <- reactiveValues()
+ for (i in r_info_elements) {
+ r_info[[i]] <- r_data[[i]]
+ }
+ suppressWarnings(rm(list = r_info_elements, envir = r_data))
+ r_info
+}
+
+## load for previous state if available but look in global memory first
+if (isTRUE(getOption("radiant.local")) && exists("r_data", envir = .GlobalEnv)) {
+ r_data <- if (is.list(r_data)) list2env(r_data, envir = new.env()) else r_data
+ if (exists("r_info")) {
+ r_info <- do.call(reactiveValues, r_info)
+ } else {
+ r_info <- r_info_legacy()
+ }
+ r_state <- if (exists("r_state")) r_state else list()
+ suppressWarnings(rm(r_data, r_state, r_info, envir = .GlobalEnv))
+} else if (isTRUE(getOption("radiant.local")) && !is.null(r_sessions[[r_ssuid]]$r_data)) {
+ r_data <- r_sessions[[r_ssuid]]$r_data %>%
+ {
+ if (is.list(.)) list2env(., envir = new.env()) else .
+ }
+ if (is.null(r_sessions[[r_ssuid]]$r_info)) {
+ r_info <- r_info_legacy()
+ } else {
+ r_info <- do.call(reactiveValues, r_sessions[[r_ssuid]]$r_info)
+ }
+ r_state <- r_sessions[[r_ssuid]]$r_state
+} else if (file.exists(paste0("~/.radiant.sessions/r_", r_ssuid, ".state.rda"))) {
+ ## read from file if not in global
+ fn <- paste0(normalizePath("~/.radiant.sessions"), "/r_", r_ssuid, ".state.rda")
+ rs <- new.env(emptyenv())
+ try(load(fn, envir = rs), silent = TRUE)
+ if (inherits(rs, "try-error")) {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ r_state <- list()
+ } else {
+ if (length(rs$r_data) == 0) {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ } else {
+ r_data <- rs$r_data %>%
+ {
+ if (is.list(.)) list2env(., envir = new.env()) else .
+ }
+ if (is.null(rs$r_info)) {
+ r_info <- r_info_legacy()
+ } else {
+ r_info <- do.call(reactiveValues, rs$r_info)
+ }
+ }
+ if (length(rs$r_state) == 0) {
+ r_state <- list()
+ } else {
+ r_state <- rs$r_state
+ }
+ }
+
+ unlink(fn, force = TRUE)
+ rm(rs)
+} else if (isTRUE(getOption("radiant.local")) && file.exists(paste0("~/.radiant.sessions/r_", mrsf, ".state.rda"))) {
+ ## restore from local folder but assign new ssuid
+ fn <- paste0(normalizePath("~/.radiant.sessions"), "/r_", mrsf, ".state.rda")
+ rs <- new.env(emptyenv())
+ try(load(fn, envir = rs), silent = TRUE)
+ if (inherits(rs, "try-error")) {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ r_state <- list()
+ } else {
+ if (length(rs$r_data) == 0) {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ } else {
+ r_data <- rs$r_data %>%
+ {
+ if (is.list(.)) list2env(., envir = new.env()) else .
+ }
+ r_info <- if (length(rs$r_info) == 0) {
+ r_info <- r_info_legacy()
+ } else {
+ do.call(reactiveValues, rs$r_info)
+ }
+ }
+ r_state <- if (length(rs$r_state) == 0) list() else rs$r_state
+ }
+
+ ## don't navigate to same tab in case the app locks again
+ r_state$nav_radiant <- NULL
+ unlink(fn, force = TRUE)
+ rm(rs)
+} else {
+ r_data <- new.env()
+ r_info <- init_data(env = r_data)
+ r_state <- list()
+}
+
+isolate({
+ for (ds in r_info[["datasetlist"]]) {
+ if (exists(ds, envir = r_data) && !bindingIsActive(as.symbol(ds), env = r_data)) {
+ shiny::makeReactiveBinding(ds, env = r_data)
+ }
+ }
+ for (dt in r_info[["dtree_list"]]) {
+ if (exists(dt, envir = r_data)) {
+ r_data[[dt]] <- add_class(r_data[[dt]], "dtree")
+ if (!bindingIsActive(as.symbol(dt), env = r_data)) {
+ shiny::makeReactiveBinding(dt, env = r_data)
+ }
+ }
+ }
+})
+
+## legacy, to deal with state files created before
+## Report > Rmd and Report > R name change
+if (isTRUE(r_state$nav_radiant == "Code")) {
+ r_state$nav_radiant <- "R"
+} else if (isTRUE(r_state$nav_radiant == "Report")) {
+ r_state$nav_radiant <- "Rmd"
+}
+
+## legacy, to deal with radio buttons that were in Data > Pivot
+if (!is.null(r_state$pvt_type)) {
+ if (isTRUE(r_state$pvt_type == "fill")) {
+ r_state$pvt_type <- TRUE
+ } else {
+ r_state$pvt_type <- FALSE
+ }
+}
+
+## legacy, to deal with state files created before
+## name change to rmd_edit
+if (!is.null(r_state$rmd_report) && is.null(r_state$rmd_edit)) {
+ r_state$rmd_edit <- r_state$rmd_report
+ r_state$rmd_report <- NULL
+}
+
+if (length(r_state$rmd_edit) > 0) {
+ r_state$rmd_edit <- r_state$rmd_edit %>% radiant.data::fix_smart()
+}
+
+## legacy, to deal with state files created before
+## name change to rmd_edit
+if (!is.null(r_state$rcode_edit) && is.null(r_state$r_edit)) {
+ r_state$r_edit <- r_state$rcode_edit
+ r_state$rcode_edit <- NULL
+}
+
+## parse the url and use updateTabsetPanel to navigate to the desired tab
+## currently only works with a new or refreshed session
+observeEvent(session$clientData$url_search, {
+ url_query <- parseQueryString(session$clientData$url_search)
+ if ("url" %in% names(url_query)) {
+ r_info[["url"]] <- url_query$url
+ } else if (is.empty(r_info[["url"]])) {
+ return()
+ }
+
+ ## create an observer and suspend when done
+ url_observe <- observe({
+ if (is.null(input$dataset)) {
+ return()
+ }
+ url <- getOption("radiant.url.patterns")[[r_info[["url"]]]]
+ if (is.null(url)) {
+ ## if pattern not found suspend observer
+ url_observe$suspend()
+ return()
+ }
+ ## move through the url
+ for (u in names(url)) {
+ if (is.null(input[[u]])) {
+ return()
+ }
+ if (input[[u]] != url[[u]]) {
+ updateTabsetPanel(session, u, selected = url[[u]])
+ }
+ if (names(tail(url, 1)) == u) url_observe$suspend()
+ }
+ })
+})
+
+## keeping track of the main tab we are on
+observeEvent(input$nav_radiant, {
+ if (!input$nav_radiant %in% c("Refresh", "Stop")) {
+ r_info[["nav_radiant"]] <- input$nav_radiant
+ }
+})
+
+## Jump to the page you were on
+## only goes two layers deep at this point
+if (!is.null(r_state$nav_radiant)) {
+ ## don't return-to-the-spot if that was quit or stop
+ if (r_state$nav_radiant %in% c("Refresh", "Stop")) {
+ return()
+ }
+
+ ## naming the observer so we can suspend it when done
+ nav_observe <- observe({
+ ## needed to avoid errors when no data is available yet
+ if (is.null(input$dataset)) {
+ return()
+ }
+ updateTabsetPanel(session, "nav_radiant", selected = r_state$nav_radiant)
+
+ ## check if shiny set the main tab to the desired value
+ if (is.null(input$nav_radiant)) {
+ return()
+ }
+ if (input$nav_radiant != r_state$nav_radiant) {
+ return()
+ }
+ nav_radiant_tab <- getOption("radiant.url.list")[[r_state$nav_radiant]] %>%
+ names()
+
+ if (!is.null(nav_radiant_tab) && !is.null(r_state[[nav_radiant_tab]])) {
+ updateTabsetPanel(session, nav_radiant_tab, selected = r_state[[nav_radiant_tab]])
+ }
+
+ ## once you arrive at the desired tab suspend the observer
+ nav_observe$suspend()
+ })
+}
+
+isolate({
+ if (is.null(r_info[["plot_height"]])) r_info[["plot_height"]] <- 650
+ if (is.null(r_info[["plot_width"]])) r_info[["plot_width"]] <- 650
+})
+
+if (getOption("radiant.from.package", default = TRUE)) {
+ ## launch using installed radiant.data package
+ # radiant.data::copy_all("radiant.data")
+ cat("\nGetting radiant.data from package ...\n")
+} else {
+ ## for shiny-server and development
+ for (file in list.files("../../R", pattern = "\\.(r|R)$", full.names = TRUE)) {
+ source(file, encoding = getOption("radiant.encoding", "UTF-8"), local = TRUE)
+ }
+ cat("\nGetting radiant.data from source ...\n")
+}
+
+## Getting screen width ...
+## https://github.com/rstudio/rstudio/issues/1870
+## https://community.rstudio.com/t/rstudio-resets-width-option-when-running-shiny-app-in-viewer/3661
+observeEvent(input$get_screen_width, {
+ if (getOption("width", default = 250) != 250) options(width = 250)
+})
+
+
+radiant.data::copy_from(radiant.data, register, deregister)
diff --git a/radiant.data/inst/app/radiant.R b/radiant.data/inst/app/radiant.R
new file mode 100644
index 0000000000000000000000000000000000000000..202656858a478f849a2e50d7bb1603a3c15069f6
--- /dev/null
+++ b/radiant.data/inst/app/radiant.R
@@ -0,0 +1,1034 @@
+################################################################################
+## function to save app state on refresh or crash
+################################################################################
+
+## drop NULLs in list
+toList <- function(x) reactiveValuesToList(x) %>% .[!sapply(., is.null)]
+
+## from https://gist.github.com/hadley/5434786
+env2list <- function(x) mget(ls(x), x)
+
+is_active <- function(env = r_data) {
+ sapply(ls(envir = env), function(x) bindingIsActive(as.symbol(x), env = env))
+}
+
+## remove non-active bindings
+rem_non_active <- function(env = r_data) {
+ iact <- is_active(env = r_data)
+ rm(list = names(iact)[!iact], envir = env)
+}
+
+active2list <- function(env = r_data) {
+ iact <- is_active(env = r_data) %>% (function(x) names(x)[x])
+ if (length(iact) > 0) {
+ mget(iact, env)
+ } else {
+ list()
+ }
+}
+
+## deal with https://github.com/rstudio/shiny/issues/2065
+MRB <- function(x, env = parent.frame(), init = FALSE) {
+ if (exists(x, envir = env)) {
+ ## if the object exists and has a binding, don't do anything
+ if (!bindingIsActive(as.symbol(x), env = env)) {
+ shiny::makeReactiveBinding(x, env = env)
+ }
+ } else if (init) {
+ ## initialize a binding (and value) if object doesn't exist yet
+ shiny::makeReactiveBinding(x, env = env)
+ }
+}
+
+saveSession <- function(session = session, timestamp = FALSE, path = "~/.radiant.sessions") {
+ if (!exists("r_sessions")) {
+ return()
+ }
+ if (!dir.exists(path)) dir.create(path)
+ isolate({
+ LiveInputs <- toList(input)
+ r_state[names(LiveInputs)] <- LiveInputs
+
+ ## removing the non-active bindings
+ rem_non_active()
+
+ r_data <- env2list(r_data)
+ r_info <- toList(r_info)
+
+ r_sessions[[r_ssuid]] <- list(
+ r_data = r_data,
+ r_info = r_info,
+ r_state = r_state,
+ timestamp = Sys.time()
+ )
+
+ ## saving session information to state file
+ if (timestamp) {
+ fn <- paste0(normalizePath(path), "/r_", r_ssuid, "-", gsub("( |:)", "-", Sys.time()), ".state.rda")
+ } else {
+ fn <- paste0(normalizePath(path), "/r_", r_ssuid, ".state.rda")
+ }
+ save(r_data, r_info, r_state, file = fn)
+ })
+}
+
+observeEvent(input$refresh_radiant, {
+ if (isTRUE(getOption("radiant.local"))) {
+ fn <- normalizePath("~/.radiant.sessions")
+ file.remove(list.files(fn, full.names = TRUE))
+ } else {
+ fn <- paste0(normalizePath("~/.radiant.sessions"), "/r_", r_ssuid, ".state.rda")
+ if (file.exists(fn)) unlink(fn, force = TRUE)
+ }
+
+ try(r_ssuid <- NULL, silent = TRUE)
+})
+
+saveStateOnRefresh <- function(session = session) {
+ session$onSessionEnded(function() {
+ isolate({
+ url_query <- parseQueryString(session$clientData$url_search)
+ if (not_pressed(input$refresh_radiant) &&
+ not_pressed(input$stop_radiant) &&
+ not_pressed(input$state_load) &&
+ not_pressed(input$state_upload) &&
+ !"fixed" %in% names(url_query)) {
+ saveSession(session)
+ } else {
+ if (not_pressed(input$state_load) && not_pressed(input$state_upload)) {
+ if (exists("r_sessions")) {
+ sshhr(try(r_sessions[[r_ssuid]] <- NULL, silent = TRUE))
+ sshhr(try(rm(r_ssuid), silent = TRUE))
+ }
+ }
+ }
+ })
+ })
+}
+
+################################################################
+## functions used across tools in radiant
+################################################################
+
+## get active dataset and apply data-filter if available
+.get_data <- reactive({
+ req(input$dataset)
+
+ filter_cmd <- input$data_filter %>%
+ gsub("\\n", "", .) %>%
+ gsub("\"", "\'", .) %>%
+ fix_smart()
+
+ arrange_cmd <- input$data_arrange
+ if (!is.empty(arrange_cmd)) {
+ arrange_cmd <- arrange_cmd %>%
+ strsplit(., split = "(&|,|\\s+)") %>%
+ unlist() %>%
+ .[!. == ""] %>%
+ paste0(collapse = ", ") %>%
+ (function(x) glue("arrange(x, {x})"))
+ }
+
+ slice_cmd <- input$data_rows
+
+ if ((is.empty(filter_cmd) && is.empty(arrange_cmd) && is.empty(slice_cmd)) || input$show_filter == FALSE) {
+ isolate(r_info[["filter_error"]] <- "")
+ } else if (grepl("([^=!<>])=([^=])", filter_cmd)) {
+ isolate(r_info[["filter_error"]] <- "Invalid filter: Never use = in a filter! Use == instead (e.g., city == 'San Diego'). Update or remove the expression")
+ } else {
+ ## %>% needed here so . will be available
+ seldat <- try(
+ r_data[[input$dataset]] %>%
+ (function(x) if (!is.empty(filter_cmd)) x %>% filter(!!rlang::parse_expr(filter_cmd)) else x) %>%
+ (function(x) if (!is.empty(arrange_cmd)) eval(parse(text = arrange_cmd)) else x) %>%
+ (function(x) if (!is.empty(slice_cmd)) x %>% slice(!!rlang::parse_expr(slice_cmd)) else x),
+ silent = TRUE
+ )
+ if (inherits(seldat, "try-error")) {
+ isolate(r_info[["filter_error"]] <- paste0("Invalid input: \"", attr(seldat, "condition")$message, "\". Update or remove the expression(x)"))
+ } else {
+ isolate(r_info[["filter_error"]] <- "")
+ if ("grouped_df" %in% class(seldat)) {
+ return(droplevels(ungroup(seldat)))
+ } else {
+ return(droplevels(seldat))
+ }
+ }
+ }
+ if ("grouped_df" %in% class(r_data[[input$dataset]])) {
+ ungroup(r_data[[input$dataset]])
+ } else {
+ r_data[[input$dataset]]
+ }
+})
+
+## using a regular function to avoid a full data copy
+.get_data_transform <- function(dataset = input$dataset) {
+ if (is.null(dataset)) {
+ return()
+ }
+ if ("grouped_df" %in% class(r_data[[dataset]])) {
+ ungroup(r_data[[dataset]])
+ } else {
+ r_data[[dataset]]
+ }
+}
+
+.get_class <- reactive({
+ get_class(.get_data())
+})
+
+groupable_vars <- reactive({
+ .get_data() %>%
+ summarise_all(
+ list(
+ ~ is.factor(.) || is.logical(.) || lubridate::is.Date(.) ||
+ is.integer(.) || is.character(.) ||
+ ((length(unique(.)) / n()) < 0.30)
+ )
+ ) %>%
+ (function(x) which(x == TRUE)) %>%
+ varnames()[.]
+})
+
+groupable_vars_nonum <- reactive({
+ .get_data() %>%
+ summarise_all(
+ list(
+ ~ is.factor(.) || is.logical(.) ||
+ lubridate::is.Date(.) || is.integer(.) ||
+ is.character(.)
+ )
+ ) %>%
+ (function(x) which(x == TRUE)) %>%
+ varnames()[.]
+})
+
+## used in compare proportions, logistic, etc.
+two_level_vars <- reactive({
+ two_levs <- function(x) {
+ if (is.factor(x)) {
+ length(levels(x))
+ } else {
+ length(unique(na.omit(x)))
+ }
+ }
+ .get_data() %>%
+ summarise_all(two_levs) %>%
+ (function(x) x == 2) %>%
+ which(.) %>%
+ varnames()[.]
+})
+
+## used in visualize - don't plot Y-variables that don't vary
+varying_vars <- reactive({
+ .get_data() %>%
+ summarise_all(does_vary) %>%
+ as.logical() %>%
+ which() %>%
+ varnames()[.]
+})
+
+## getting variable names in active dataset and their class
+varnames <- reactive({
+ var_class <- .get_class()
+ req(var_class)
+ names(var_class) %>%
+ set_names(., paste0(., " {", var_class, "}"))
+})
+
+## cleaning up the arguments for data_filter and defaults passed to report
+clean_args <- function(rep_args, rep_default = list()) {
+ if (!is.null(rep_args$data_filter)) {
+ if (rep_args$data_filter == "") {
+ rep_args$data_filter <- NULL
+ } else {
+ rep_args$data_filter %<>% gsub("\\n", "", .) %>% gsub("\"", "\'", .)
+ }
+ }
+ if (is.empty(rep_args$rows)) {
+ rep_args$rows <- NULL
+ }
+ if (is.empty(rep_args$arr)) {
+ rep_args$arr <- NULL
+ }
+
+ if (length(rep_default) == 0) rep_default[names(rep_args)] <- ""
+
+ ## removing default arguments before sending to report feature
+ for (i in names(rep_args)) {
+ if (!is.language(rep_args[[i]]) && !is.call(rep_args[[i]]) && all(is.na(rep_args[[i]]))) {
+ rep_args[[i]] <- NULL
+ next
+ }
+ if (!is.symbol(rep_default[[i]]) && !is.call(rep_default[[i]]) && all(is_not(rep_default[[i]]))) next
+ if (length(rep_args[[i]]) == length(rep_default[[i]]) && !is.name(rep_default[[i]]) && all(rep_args[[i]] == rep_default[[i]])) {
+ rep_args[[i]] <- NULL
+ }
+ }
+
+ rep_args
+}
+
+## check if a variable is null or not in the selected data.frame
+not_available <- function(x) any(is.null(x)) || (sum(x %in% varnames()) < length(x))
+
+## check if a variable is null or not in the selected data.frame
+available <- function(x) !not_available(x)
+
+## check if a button was pressed
+pressed <- function(x) !is.null(x) && (is.list(x) || x > 0)
+
+## check if a button was NOT pressed
+not_pressed <- function(x) !pressed(x)
+
+## check for duplicate entries
+has_duplicates <- function(x) length(unique(x)) < length(x)
+
+## is x some type of date variable
+is_date <- function(x) inherits(x, c("Date", "POSIXlt", "POSIXct"))
+
+## drop elements from .._args variables obtained using formals
+r_drop <- function(x, drop = c("dataset", "data_filter", "arr", "rows", "envir")) x[!x %in% drop]
+
+## show a few rows of a dataframe
+show_data_snippet <- function(dataset = input$dataset, nshow = 7, title = "", filt = "", arr = "", rows = "") {
+ if (is.character(dataset) && length(dataset) == 1) dataset <- get_data(dataset, filt = filt, arr = arr, rows = rows, na.rm = FALSE, envir = r_data)
+ nr <- nrow(dataset)
+ ## avoid slice with variables outside of the df in case a column with the same
+ ## name exists
+ dataset[1:min(nshow, nr), , drop = FALSE] %>%
+ mutate_if(is_date, as.character) %>%
+ mutate_if(is.character, list(~ strtrim(., 40))) %>%
+ xtable::xtable(.) %>%
+ print(
+ type = "html", print.results = FALSE, include.rownames = FALSE,
+ sanitize.text.function = identity,
+ html.table.attributes = "class='table table-condensed table-hover snippet'"
+ ) %>%
+ paste0(title, .) %>%
+ (function(x) if (nr <= nshow) x else paste0(x, "\n")) %>%
+ enc2utf8()
+}
+
+suggest_data <- function(text = "", df_name = "diamonds") {
+ paste0(text, "要获取示例数据集,请转到数据 > 管理,从加载数据类型'下拉菜单中选择'示例',\n然后点击'加载'按钮,选择 '", df_name, "' 数据集。")
+}
+
+## function written by @wch https://github.com/rstudio/shiny/issues/781#issuecomment-87135411
+capture_plot <- function(expr, env = parent.frame()) {
+ structure(
+ list(expr = substitute(expr), env = env),
+ class = "capture_plot"
+ )
+}
+
+## function written by @wch https://github.com/rstudio/shiny/issues/781#issuecomment-87135411
+print.capture_plot <- function(x, ...) {
+ eval(x$expr, x$env)
+}
+
+################################################################
+## functions used to create Shiny in and outputs
+################################################################
+
+## textarea where the return key submits the content
+returnTextAreaInput <- function(inputId, label = NULL, rows = 2,
+ placeholder = NULL, resize = "vertical",
+ value = "") {
+ ## avoid all sorts of 'helpful' behavior from your browser
+ ## see https://stackoverflow.com/a/35514029/1974918
+ tagList(
+ tags$div(
+ # using containing element based on
+ # https://github.com/niklasvh/html2canvas/issues/2008#issuecomment-1445503369
+ tags$label(label, `for` = inputId), br(),
+ tags$textarea(
+ value,
+ id = inputId,
+ type = "text",
+ rows = rows,
+ placeholder = placeholder,
+ resize = resize,
+ autocomplete = "off",
+ autocorrect = "off",
+ autocapitalize = "off",
+ spellcheck = "false",
+ class = "returnTextArea form-control"
+ )
+ )
+ )
+}
+
+## from https://github.com/rstudio/shiny/blob/master/R/utils.R
+`%AND%` <- function(x, y) {
+ if (!all(is.null(x)) && !all(is.na(x))) {
+ if (!all(is.null(y)) && !all(is.na(y))) {
+ return(y)
+ }
+ }
+ return(NULL)
+}
+
+## using a custom version of textInput to avoid browser "smartness"
+textInput <- function(inputId, label, value = "", width = NULL,
+ placeholder = NULL, autocomplete = "off",
+ autocorrect = "off", autocapitalize = "off",
+ spellcheck = "false", ...) {
+ value <- restoreInput(id = inputId, default = value)
+ div(
+ class = "form-group shiny-input-container",
+ style = if (!is.null(width)) paste0("width: ", validateCssUnit(width), ";"),
+ label %AND% tags$label(label, `for` = inputId),
+ tags$input(
+ id = inputId,
+ type = "text",
+ class = "form-control",
+ value = value,
+ placeholder = placeholder,
+ autocomplete = autocomplete,
+ autocorrect = autocorrect,
+ autocapitalize = autocapitalize,
+ spellcheck = spellcheck,
+ ...
+ )
+ )
+}
+
+## using a custom version of textAreaInput to avoid browser "smartness"
+textAreaInput <- function(inputId, label, value = "", width = NULL,
+ height = NULL, cols = NULL, rows = NULL,
+ placeholder = NULL, resize = NULL,
+ autocomplete = "off", autocorrect = "off",
+ autocapitalize = "off", spellcheck = "true",
+ ...) {
+ value <- restoreInput(id = inputId, default = value)
+ if (!is.null(resize)) {
+ resize <- match.arg(
+ resize,
+ c("both", "none", "vertical", "horizontal")
+ )
+ }
+ style <- paste(if (!is.null(width)) {
+ paste0("width: ", validateCssUnit(width), ";")
+ }, if (!is.null(height)) {
+ paste0("height: ", validateCssUnit(height), ";")
+ }, if (!is.null(resize)) {
+ paste0("resize: ", resize, ";")
+ })
+ if (length(style) == 0) {
+ style <- NULL
+ }
+ div(
+ class = "form-group shiny-input-container",
+ label %AND% tags$label(label, `for` = inputId),
+ tags$textarea(
+ id = inputId,
+ class = "form-control",
+ placeholder = placeholder,
+ style = style,
+ rows = rows,
+ cols = cols,
+ autocomplete = autocomplete,
+ autocorrect = autocorrect,
+ autocapitalize = autocapitalize,
+ spellcheck = spellcheck,
+ ...,
+ value
+ )
+ )
+}
+
+## avoid all sorts of 'helpful' behavior from your browser
+## based on https://stackoverflow.com/a/35514029/1974918
+returnTextInput <- function(inputId, label = NULL,
+ placeholder = NULL, value = "") {
+ tagList(
+ tags$label(label, `for` = inputId),
+ tags$input(
+ id = inputId,
+ type = "text",
+ value = value,
+ placeholder = placeholder,
+ autocomplete = "off",
+ autocorrect = "off",
+ autocapitalize = "off",
+ spellcheck = "false",
+ class = "returnTextInput form-control"
+ )
+ )
+}
+
+if (getOption("radiant.shinyFiles", FALSE)) {
+ download_link <- function(id) {
+ uiOutput(paste0("ui_", id))
+ }
+
+ download_button <- function(id, ...) {
+ uiOutput(paste0("ui_", id))
+ }
+
+ download_handler <- function(id, label = "", fun = id, fn, type = "csv", caption = "Save to csv",
+ class = "", ic = "download", btn = "link", onclick = "function() none;", ...) {
+ ## create observer
+ shinyFiles::shinyFileSave(input, id, roots = sf_volumes, session = session)
+
+ ## create renderUI
+ if (btn == "link") {
+ output[[paste0("ui_", id)]] <- renderUI({
+ if (is.function(fn)) fn <- fn()
+ if (is.function(type)) type <- type()
+ shinyFiles::shinySaveLink(
+ id, label, caption,
+ filename = fn, filetype = type,
+ class = "alignright", icon = icon(ic, verify_fa = FALSE), onclick = onclick
+ )
+ })
+ } else {
+ output[[paste0("ui_", id)]] <- renderUI({
+ if (is.function(fn)) fn <- fn()
+ if (is.function(type)) type <- type()
+ shinyFiles::shinySaveButton(
+ id, label, caption,
+ filename = fn, filetype = type,
+ class = class, icon = icon("download", verify_fa = FALSE), onclick = onclick
+ )
+ })
+ }
+
+ observeEvent(input[[id]], {
+ if (is.integer(input[[id]])) {
+ return()
+ }
+ path <- shinyFiles::parseSavePath(sf_volumes, input[[id]])
+ if (!inherits(path, "try-error") && !is.empty(path$datapath)) {
+ fun(path$datapath, ...)
+ }
+ })
+ }
+} else {
+ download_link <- function(id, ...) {
+ downloadLink(id, "", class = "fa fa-download alignright", ...)
+ }
+ download_button <- function(id, label = "Save", ic = "download", class = "", ...) {
+ downloadButton(id, label, class = class, ...)
+ }
+ download_handler <- function(id, label = "", fun = id, fn, type = "csv", caption = "Save to csv",
+ class = "", ic = "download", btn = "link", ...) {
+ output[[id]] <- downloadHandler(
+ filename = function() {
+ if (is.function(fn)) fn <- fn()
+ if (is.function(type)) type <- type()
+ paste0(fn, ".", type)
+ },
+ content = function(path) {
+ fun(path, ...)
+ }
+ )
+ }
+}
+
+plot_width <- function() {
+ if (is.null(input$viz_plot_width)) r_info[["plot_width"]] else input$viz_plot_width
+}
+
+plot_height <- function() {
+ if (is.null(input$viz_plot_height)) r_info[["plot_height"]] else input$viz_plot_height
+}
+
+download_handler_plot <- function(path, plot, width = plot_width, height = plot_height) {
+ plot <- try(plot(), silent = TRUE)
+ if (inherits(plot, "try-error") || is.character(plot) || is.null(plot)) {
+ plot <- ggplot() +
+ labs(title = "Plot not available")
+ inp <- c(500, 100, 96)
+ } else {
+ inp <- 5 * c(width(), height(), 96)
+ }
+
+ png(file = path, width = inp[1], height = inp[2], res = inp[3])
+ print(plot)
+ dev.off()
+}
+
+## fun_name is a string of the main function name
+## rfun_name is a string of the reactive wrapper that calls the main function
+## out_name is the name of the output, set to fun_name by default
+register_print_output <- function(fun_name, rfun_name, out_name = fun_name) {
+ ## Generate output for the summary tab
+ output[[out_name]] <- renderPrint({
+ ## when no analysis was conducted (e.g., no variables selected)
+ fun <- get(rfun_name)()
+ if (is.character(fun)) {
+ cat(fun, "\n")
+ } else {
+ rm(fun)
+ }
+ })
+ return(invisible())
+}
+
+## fun_name is a string of the main function name
+## rfun_name is a string of the reactive wrapper that calls the main function
+## out_name is the name of the output, set to fun_name by default
+register_plot_output <- function(fun_name, rfun_name, out_name = fun_name,
+ width_fun = "plot_width", height_fun = "plot_height") {
+ ## Generate output for the plots tab
+ output[[out_name]] <- renderPlot(
+ {
+ ## when no analysis was conducted (e.g., no variables selected)
+ p <- get(rfun_name)()
+ if (is_not(p) || is.empty(p)) p <- "Nothing to plot ...\nSelect plots to show or re-run the calculations"
+ if (is.character(p)) {
+ plot(
+ x = 1, type = "n", main = paste0("\n\n\n\n\n\n\n\n", p),
+ axes = FALSE, xlab = "", ylab = "", cex.main = .9
+ )
+ } else {
+ print(p)
+ }
+ },
+ width = get(width_fun),
+ height = get(height_fun),
+ res = 96
+ )
+
+ return(invisible())
+}
+
+stat_tab_panel <- function(menu, tool, tool_ui, output_panels,
+ data = input$dataset) {
+ sidebarLayout(
+ sidebarPanel(
+ wellPanel(
+ HTML(paste("")), + HTML(paste("
")), + if (!is.null(data)) { + HTML(paste("")) + } + ), + uiOutput(tool_ui) + ), + mainPanel( + output_panels + ) + ) +} + +################################################################ +## functions used for app help +################################################################ +help_modal <- function(modal_title, link, help_file, + author = "Vincent Nijs", + year = lubridate::year(lubridate::now()), + lic = "by-nc-sa") { + sprintf( + " + ", + link, link, link, modal_title, help_file, author, year, lic, lic, link + ) %>% + enc2utf8() %>% + HTML() +} + +help_and_report <- function(modal_title, fun_name, help_file, + author = "Vincent Nijs", + year = lubridate::year(lubridate::now()), + lic = "by-nc-sa") { + sprintf( + " + + + + ", + fun_name, fun_name, fun_name, modal_title, help_file, author, year, lic, lic, fun_name, fun_name, fun_name, fun_name, fun_name + ) %>% + enc2utf8() %>% + HTML() %>% + withMathJax() +} + +## function to render .md files to html +inclMD <- function(path) { + paste(readLines(path, warn = FALSE), collapse = "\n") %>% + markdown::mark_html(text = ., template = FALSE, meta = list(css = ""), output = FALSE) +} + +## function to render .Rmd files to html +inclRmd <- function(path) { + paste(readLines(path, warn = FALSE), collapse = "\n") %>% + knitr::knit2html( + text = ., template = FALSE, quiet = TRUE, + envir = r_data, meta = list(css = ""), output = FALSE + ) %>% + HTML() %>% + withMathJax() +} + +## capture the state of a dt table +dt_state <- function(fun, vars = "", tabfilt = "", tabsort = "", nr = 0) { + ## global search + search <- input[[paste0(fun, "_state")]]$search$search + if (is.null(search)) search <- "" + + ## table ordering + order <- input[[paste0(fun, "_state")]]$order + if (length(order) == 0) { + order <- "NULL" + } else { + order <- list(order) + } + + ## column filters, gsub needed for factors + sc <- input[[paste0(fun, "_search_columns")]] %>% gsub("\\\"", "'", .) + sci <- which(sc != "") + nr_sc <- length(sci) + if (nr_sc > 0) { + sc <- list(lapply(sci, function(i) list(i, sc[i]))) + } else if (nr_sc == 0) { + sc <- "NULL" + } + + dat <- get(paste0(".", fun))()$tab %>% + (function(x) { + nr <<- nrow(x) + x[1, , drop = FALSE] + }) + + if (order != "NULL" || sc != "NULL") { + ## get variable class and name + gc <- get_class(dat) %>% + (function(x) if (is.empty(vars[1])) x else x[vars]) + cn <- names(gc) + + if (length(cn) > 0) { + if (order != "NULL") { + tabsort <- c() + for (i in order[[1]]) { + cname <- cn[i[[1]] + 1] %>% gsub("^\\s+|\\s+$", "", .) + if (grepl("[^0-9a-zA-Z_\\.]", cname) || grepl("^[0-9]", cname)) { + cname <- paste0("`", cname, "`") + } + if (i[[2]] == "desc") cname <- paste0("desc(", cname, ")") + tabsort <- c(tabsort, cname) + } + tabsort <- paste0(tabsort, collapse = ", ") + } + + if (sc != "NULL") { + tabfilt <- c() + for (i in sc[[1]]) { + cname <- cn[i[[1]]] + type <- gc[cname] + if (type == "factor") { + cname <- paste0(cname, " %in% ", sub("\\[", "c(", i[[2]]) %>% sub("\\]", ")", .)) + } else if (type %in% c("numeric", "integer", "ts")) { + bnd <- strsplit(i[[2]], "...", fixed = TRUE)[[1]] + cname <- paste0(cname, " >= ", bnd[1], " & ", cname, " <= ", bnd[2]) %>% gsub(" ", " ", .) + } else if (type %in% c("date", "period")) { + bnd <- strsplit(i[[2]], "...", fixed = TRUE)[[1]] %>% gsub(" ", "", .) + cname <- paste0(cname, " >= '", bnd[1], "' & ", cname, " <= '", bnd[2], "'") %>% gsub(" ", " ", .) + } else if (type == "character") { + cname <- paste0("grepl('", i[[2]], "', ", cname, ", ignore.case = TRUE)") + } else if (type == "logical") { + cname <- paste0(cname, " == ", toupper(sub("\\['(true|false)'\\]", "\\1", i[[2]]))) + } else { + message("Variable ", cname, " has type ", type, ". This type is not currently supported to generate code for Report > Rmd or Report > R") + next + } + tabfilt <- c(tabfilt, cname) + } + tabfilt <- paste0(tabfilt, collapse = " & ") + } + } + } + + list(search = search, order = order, sc = sc, tabsort = tabsort, tabfilt = tabfilt, nr = nr) +} + +## use the value in the input list if available and update r_state +state_init <- function(var, init = "", na.rm = TRUE) { + isolate({ + ivar <- input[[var]] + if (var %in% names(input) || length(ivar) > 0) { + ivar <- input[[var]] + if ((na.rm && is.empty(ivar)) || length(ivar) == 0) { + r_state[[var]] <<- NULL + } + } else { + ivar <- .state_init(var, init, na.rm) + } + ivar + }) +} + +## need a separate function for checkboxGroupInputs +state_group <- function(var, init = "") { + isolate({ + ivar <- input[[var]] + if (var %in% names(input) || length(ivar) > 0) { + ivar <- input[[var]] + if (is.empty(ivar)) r_state[[var]] <<- NULL + } else { + ivar <- .state_init(var, init) + r_state[[var]] <<- NULL ## line that differs for CBG inputs + } + ivar + }) +} + +.state_init <- function(var, init = "", na.rm = TRUE) { + rs <- r_state[[var]] + if ((na.rm && is.empty(rs)) || length(rs) == 0) init else rs +} + +state_single <- function(var, vals, init = character(0)) { + isolate({ + ivar <- input[[var]] + if (var %in% names(input) && is.null(ivar)) { + r_state[[var]] <<- NULL + ivar + } else if (available(ivar) && all(ivar %in% vals)) { + if (length(ivar) > 0) r_state[[var]] <<- ivar + ivar + } else if (available(ivar) && any(ivar %in% vals)) { + ivar[ivar %in% vals] + } else { + if (length(ivar) > 0 && all(ivar %in% c("None", "none", ".", ""))) { + r_state[[var]] <<- ivar + } + .state_single(var, vals, init = init) + } + }) +} + +.state_single <- function(var, vals, init = character(0)) { + rs <- r_state[[var]] + if (is.empty(rs)) init else vals[vals == rs] +} + +state_multiple <- function(var, vals, init = character(0)) { + isolate({ + ivar <- input[[var]] + if (var %in% names(input) && is.null(ivar)) { + r_state[[var]] <<- NULL + ivar + } else if (available(ivar) && all(ivar %in% vals)) { + if (length(ivar) > 0) r_state[[var]] <<- ivar + ivar + } else if (available(ivar) && any(ivar %in% vals)) { + ivar[ivar %in% vals] + } else { + if (length(ivar) > 0 && all(ivar %in% c("None", "none", ".", ""))) { + r_state[[var]] <<- ivar + } + .state_multiple(var, vals, init = init) + } + }) +} + +.state_multiple <- function(var, vals, init = character(0)) { + rs <- r_state[[var]] + r_state[[var]] <<- NULL + + ## "a" %in% character(0) --> FALSE, letters[FALSE] --> character(0) + if (is.empty(rs)) vals[vals %in% init] else vals[vals %in% rs] +} + +## cat to file +## use with tail -f ~/r_cat.txt in a terminal +# cf <- function(...) { +# cat(paste0("\n--- called from: ", environmentName(parent.frame()), " (", lubridate::now(), ")\n"), file = "~/r_cat.txt", append = TRUE) +# out <- paste0(capture.output(...), collapse = "\n") +# cat("--\n", out, "\n--", sep = "\n", file = "~/r_cat.txt", append = TRUE) +# } + +## autosave option +## provide a list with (1) the save interval in minutes, (2) the total duration in minutes, and (3) the path to use +# options(radiant.autosave = list(1, 5, "~/.radiant.sessions")) +# options(radiant.autosave = list(.1, 1, "~/Desktop/radiant.sessions")) +# options(radiant.autosave = list(10, 180, "~/Desktop/radiant.sessions")) +if (length(getOption("radiant.autosave", default = NULL)) > 0) { + start_time <- Sys.time() + interval <- getOption("radiant.autosave")[[1]] * 60000 + max_duration <- getOption("radiant.autosave")[[2]] + autosave_path <- getOption("radiant.autosave")[[3]] + autosave_path <- ifelse(length(autosave_path) == 0, "~/.radiant.sessions", autosave_path) + autosave_poll <- reactivePoll( + interval, + session, + checkFunc = function() { + curr_time <- Sys.time() + diff_time <- difftime(curr_time, start_time, units = "mins") + if (diff_time < max_duration) { + saveSession(session, timestamp = TRUE, autosave_path) + options(radiant.autosave = list(interval, max_duration - diff_time, autosave_path)) + message("Radiant state was auto-saved at ", curr_time) + } else { + if (length(getOption("radiant.autosave", default = NULL)) > 0) { + showModal( + modalDialog( + title = "Radiant state autosave", + span(glue("The autosave feature has been turned off. Time to save and submit your work by clicking + on the 'Save' icon in the navigation bar and then on 'Save radiant state file'. To clean the + state files that were auto-saved, run the following command from the R(studio) console: + unlink('{autosave_path}/*.state.rda', force = TRUE)")), + footer = modalButton("OK"), + size = "m", + easyClose = TRUE + ) + ) + options(radiant.autosave = NULL) + } + } + }, + valueFunc = function() { + return() + } + ) +} + +## update "run" button when relevant inputs are changed +run_refresh <- function(args, pre, init = "evar", tabs = "", + label = "Estimate model", relabel = label, + inputs = NULL, data = TRUE) { + observe({ + ## dep on most inputs + if (data) { + input$data_filter + input$data_arrange + input$data_rows + input$show_filter + } + sapply(r_drop(names(args)), function(x) input[[paste0(pre, "_", x)]]) + + ## adding dependence in more inputs + if (length(inputs) > 0) { + sapply(inputs, function(x) input[[paste0(pre, "_", x)]]) + } + + run <- isolate(input[[paste0(pre, "_run")]]) %>% pressed() + check_null <- function(init) { + all(sapply(init, function(x) is.null(input[[paste0(pre, "_", x)]]))) + } + if (isTRUE(check_null(init))) { + if (!is.empty(tabs)) { + updateTabsetPanel(session, paste0(tabs, " "), selected = "Summary") + } + updateActionButton(session, paste0(pre, "_run"), label, icon = icon("play", verify_fa = FALSE)) + } else if (run) { + updateActionButton(session, paste0(pre, "_run"), relabel, icon = icon("sync", class = "fa-spin", verify_fa = FALSE)) + } else { + updateActionButton(session, paste0(pre, "_run"), label, icon = icon("play", verify_fa = FALSE)) + } + }) + + observeEvent(input[[paste0(pre, "_run")]], { + updateActionButton(session, paste0(pre, "_run"), label, icon = icon("play", verify_fa = FALSE)) + }) +} + +radiant_screenshot_modal <- function(report_on = "") { + add_button <- function() { + if (is.empty(report_on)) { + "" + } else { + actionButton(report_on, i18n$t("Report"), icon = icon("edit", verify_fa = FALSE), class = "btn-success") + } + } + showModal( + modalDialog( + title = i18n$t("Radiant screenshot"), + span(shiny::tags$div(id = "screenshot_preview")), + span(HTML("要在报告中包含截图,请先点击保存按钮将截图保存到磁盘。然后点击报告按钮,将截图引用插入到报告 > Rmd中。")), + footer = tagList( + tags$table( + tags$td(download_button("screenshot_save", i18n$t("Save"), ic = "download")), + tags$td(add_button()), + tags$td(modalButton(i18n$t("Cancel"))), + align = "right" + ) + ), + size = "l", + easyClose = TRUE + ) + ) +} + +observeEvent(input$screenshot_link, { + radiant_screenshot_modal() +}) + +render_screenshot <- function() { + plt <- sub("data:.+base64,", "", input$img_src) + png::readPNG(base64enc::base64decode(what = plt)) +} + +download_handler_screenshot <- function(path, plot, ...) { + plot <- try(plot(), silent = TRUE) + if (inherits(plot, "try-error") || is.character(plot) || is.null(plot)) { + plot <- ggplot() + + labs(title = "Plot not available") + png(file = path, width = 500, height = 100, res = 96) + print(plot) + dev.off() + } else { + ppath <- parse_path(path, pdir = getOption("radiant.launch_dir", find_home()), mess = FALSE) + # r_info[["latest_screenshot"]] <- glue("") + # r_info[["latest_screenshot"]] <- glue("
Click to show screenshot
\nClick to show screenshot with Radiant settings to generate output shown below
\n\n\n diff --git a/radiant.data/inst/app/tools/app/help.R b/radiant.data/inst/app/tools/app/help.R
new file mode 100644
index 0000000000000000000000000000000000000000..0fcac28c923a08c1381bc572c9c56fe32643ea97
--- /dev/null
+++ b/radiant.data/inst/app/tools/app/help.R
@@ -0,0 +1,138 @@
+#######################################
+## Other elements in help menu
+#######################################
+output$help_videos <- renderUI({
+ file.path(getOption("radiant.path.data"), "app/tools/app/tutorials.md") %>% inclMD() %>% HTML()
+})
+
+output$help_about <- renderUI({
+ file.path(getOption("radiant.path.data"), "app/tools/app/about.md") %>% inclMD() %>% HTML()
+})
+
+output$help_text <- renderUI({
+ wellPanel(
+ HTML("Help is available on each page by clicking the icon on the bottom left of your screen.
diff --git a/radiant.data/inst/app/tools/app/help.R b/radiant.data/inst/app/tools/app/help.R
new file mode 100644
index 0000000000000000000000000000000000000000..0fcac28c923a08c1381bc572c9c56fe32643ea97
--- /dev/null
+++ b/radiant.data/inst/app/tools/app/help.R
@@ -0,0 +1,138 @@
+#######################################
+## Other elements in help menu
+#######################################
+output$help_videos <- renderUI({
+ file.path(getOption("radiant.path.data"), "app/tools/app/tutorials.md") %>% inclMD() %>% HTML()
+})
+
+output$help_about <- renderUI({
+ file.path(getOption("radiant.path.data"), "app/tools/app/about.md") %>% inclMD() %>% HTML()
+})
+
+output$help_text <- renderUI({
+ wellPanel(
+ HTML("Help is available on each page by clicking the icon on the bottom left of your screen.Versions: ", getOption("radiant.versions", default = "Unknown")) + ) +}) + +####################################### +## Main function of help menu +####################################### +append_help <- function(help_str, help_path, lic = "nc", Rmd = TRUE) { + if (length(input[[help_str]]) == 0) return() + help_block <- get(help_str) + local_hd <- help_block[which(help_block %in% input[[help_str]])] + all_help <- c() + for (i in names(local_hd)) { + all_help <- paste( + all_help, paste0("
", i, "
"), + inclRmd(file.path(help_path, local_hd[i])), + sep = "\n" + ) + } + mathjax_script <- ifelse(Rmd, "", "") + + if (lic == "nc") { + cc <- getOption("radiant.help.cc", default = "") + } else { + cc <- getOption("radiant.help.cc.sa", default = "") + } + + ## remove ` from report.md + paste( + gsub("(\"> )`", "\\1", all_help) %>% + gsub("`( )", "\\1", .), + "\n", mathjax_script, "\n", cc + ) %>% HTML() +} + +help_switch <- function(help_all, help_str, help_on = TRUE) { + if (is.null(help_all) || help_all == 0) return() + help_choices <- help_init <- get(help_str) + init <- "" + if (help_on) init <- help_init + updateCheckboxGroupInput( + session, help_str, + label = NULL, + choices = help_choices, + selected = init, inline = TRUE + ) +} + +help_data <- c( + "Manage" = "manage.md", "View" = "view.md", "Visualize" = "visualize.md", + "Pivot" = "pivotr.md", "Explore" = "explore.md", "Transform" = "transform.md", + "Combine" = "combine.md", "Report > Rmd" = "report_rmd.md", "Report > R" = "report_r.md" +) +output$help_data <- reactive( + append_help("help_data", file.path(getOption("radiant.path.data"), "app/tools/help/")) +) + +observeEvent(input$help_data_all, { + help_switch(input$help_data_all, "help_data") +}) +observeEvent(input$help_data_none, { + help_switch(input$help_data_none, "help_data", help_on = FALSE) +}) + +help_data_panel <- + wellPanel( + HTML( + "" + ), + checkboxGroupInput("help_data", NULL, help_data, selected = state_group("help_data"), inline = TRUE) + ) + +output$help_data_ui <- renderUI({ + sidebarLayout( + sidebarPanel( + help_data_panel, + uiOutput("help_text"), + width = 3 + ), + mainPanel( + HTML(paste0("Select help files to show and search
")), + htmlOutput("help_data") + ) + ) +}) + +observeEvent(input$help_keyboard, { + showModal( + modalDialog( + title = i18n$t("Keyboard shortcuts"), + h4(i18n$t("General")), + ## based on https://github.com/swarm-lab/editR/blob/master/inst/app/bits/keyboard.R + withTags( + table(style = "width: 80%; margin-left: 10%;", + tr(class = "border_bottom", + td(b(i18n$t("Function"))), td(b("Mac")), td(b("Windows & Linux"))), + tr(class = "padding_top", + td(i18n$t("Save state")), td("Shift-CMD-s"), td("Shift-CTRL-s")), + tr(class = "padding_top", + td(i18n$t("Open state")), td("Shift-CMD-o"), td("Shift-CTRL-o")), + tr(class = "border_bottom padding_bottom", + td(i18n$t("Show help")), td("F1"), td("F1")), + tr(class = "border_bottom padding_bottom", + td(i18n$t("Generate screenshot")), td("CMD-p"), td("CTRL-p")), + tr(class = "border_bottom padding_bottom", + td(i18n$t("Generate code")), td("ALT-return"), td("ALT-return")), + tr(class = "border_bottom padding_bottom", + td(i18n$t("Estimate/Run (green button)")), td("CMD-enter"), td("CTRL-enter")), + tr(class = "border_bottom padding_bottom", + td(i18n$t("Save (blue button)")), td("CMD-s"), td("CTRL-s")), + tr(class = "border_bottom padding_bottom", + td(i18n$t("Download (blue icon)")), td("CMD-s"), td("CTRL-s")), + tr(class = "border_bottom padding_bottom", + td(i18n$t("Load (blue button)")), td("CMD-o"), td("CTRL-o")) + # tr(class = "border_bottom padding_bottom", + # td("Viewer pane full screen"), td("Shift-CTRL-9"), td("Shift-CTRL-9")) + ) + ), + footer = modalButton("OK"), + size = "l", + easyClose = TRUE + ) + ) +}) diff --git a/radiant.data/inst/app/tools/app/report_funs.R b/radiant.data/inst/app/tools/app/report_funs.R new file mode 100644 index 0000000000000000000000000000000000000000..023d8f60f1ad86483b3231a5041e9897d0271212 --- /dev/null +++ b/radiant.data/inst/app/tools/app/report_funs.R @@ -0,0 +1,850 @@ +file_upload_button <- function(inputId, label = "", multiple = FALSE, + accept = NULL, buttonLabel = "Load", title = "Load data", + class = "", icn = "upload", progress = FALSE) { + if (getOption("radiant.shinyFiles", FALSE)) { + shinyFiles::shinyFileChoose( + input = input, + id = inputId, + session = session, + roots = sf_volumes, + filetype = gsub(".", "", accept, fixed = TRUE) + ) + + # actionButton(inputId, buttonLabel, icon = icon(icn), class = class) + shinyFiles::shinyFilesButton( + inputId, buttonLabel, label, + title = title, multiple = FALSE, + class = class, icon = icon(icn, verify_fa = FALSE) + ) + } else { + if (length(accept) > 0) { + accept <- paste(accept, collapse = ",") + } else { + accept <- "" + } + + if (!is.empty(label)) { + label <- paste0("") + } + + btn <- paste0(label, " + + ") + + if (progress) { + btn <- paste0(btn, "\n
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +
| Storm | +good | +female | +Marvel | +
| Mystique | +bad | +female | +Marvel | +
| Batman | +good | +male | +DC | +
| Joker | +bad | +male | +DC | +
| Catwoman | +bad | +female | +DC | +
| Hellboy | +good | +male | +Dark Horse Comics | +
| publisher | +yr_founded | +
|---|---|
| DC | +1934 | +
| Marvel | +1939 | +
| Image | +1992 | +

+ +### 内连接(超级英雄 × 出版商) + +若 x = 超级英雄数据集,y = 出版商数据集: + +> 内连接返回 x 中与 y 有匹配值的所有行,以及 x 和 y 的所有列。若 x 和 y 之间存在多个匹配,所有匹配组合都会被返回。 + +
| name | +alignment | +gender | +publisher | +yr_founded | +
|---|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +1939 | +
| Storm | +good | +female | +Marvel | +1939 | +
| Mystique | +bad | +female | +Marvel | +1939 | +
| Batman | +good | +male | +DC | +1934 | +
| Joker | +bad | +male | +DC | +1934 | +
| Catwoman | +bad | +female | +DC | +1934 | +

+ +### 左连接(超级英雄 × 出版商) + +> 左连接返回 x 的所有行,以及 x 和 y 的所有列。若 x 和 y 之间存在多个匹配,所有匹配组合都会被返回。 + +
| name | +alignment | +gender | +publisher | +yr_founded | +
|---|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +1939 | +
| Storm | +good | +female | +Marvel | +1939 | +
| Mystique | +bad | +female | +Marvel | +1939 | +
| Batman | +good | +male | +DC | +1934 | +
| Joker | +bad | +male | +DC | +1934 | +
| Catwoman | +bad | +female | +DC | +1934 | +
| Hellboy | +good | +male | +Dark Horse Comics | +NA | +

+ +### 右连接(超级英雄 × 出版商) + +> 右连接返回 y 的所有行,以及 y 和 x 的所有列。若 y 和 x 之间存在多个匹配,所有匹配组合都会被返回。 + +
| name | +alignment | +gender | +publisher | +yr_founded | +
|---|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +1939 | +
| Storm | +good | +female | +Marvel | +1939 | +
| Mystique | +bad | +female | +Marvel | +1939 | +
| Batman | +good | +male | +DC | +1934 | +
| Joker | +bad | +male | +DC | +1934 | +
| Catwoman | +bad | +female | +DC | +1934 | +
| NA | +NA | +NA | +Image | +1992 | +

+ +### 全连接(超级英雄 × 出版商) + +> 全连接合并两个数据集,保留出现在任一数据集中的行和列。 + +
| name | +alignment | +gender | +publisher | +yr_founded | +
|---|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +1939 | +
| Storm | +good | +female | +Marvel | +1939 | +
| Mystique | +bad | +female | +Marvel | +1939 | +
| Batman | +good | +male | +DC | +1934 | +
| Joker | +bad | +male | +DC | +1934 | +
| Catwoman | +bad | +female | +DC | +1934 | +
| Hellboy | +good | +male | +Dark Horse Comics | +NA | +
| NA | +NA | +NA | +Image | +1992 | +

| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +
| Storm | +good | +female | +Marvel | +
| Mystique | +bad | +female | +Marvel | +
| Batman | +good | +male | +DC | +
| Joker | +bad | +male | +DC | +
| Catwoman | +bad | +female | +DC | +
+ +### 反连接(超级英雄 × 出版商) + +> 反连接返回 x 中与 y 无匹配值的所有行,仅保留 x 中的列。 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Hellboy | +good | +male | +Dark Horse Comics | +

+ +### 数据集顺序 + +请注意,所选数据集的顺序可能会影响连接结果。如果我们如下设置 `数据> 合并` 标签页,结果如下: + +

+ +### 内连接(出版商 × 超级英雄) + +
| publisher | +yr_founded | +name | +alignment | +gender | +
|---|---|---|---|---|
| DC | +1934 | +Batman | +good | +male | +
| DC | +1934 | +Joker | +bad | +male | +
| DC | +1934 | +Catwoman | +bad | +female | +
| Marvel | +1939 | +Magneto | +bad | +male | +
| Marvel | +1939 | +Storm | +good | +female | +
| Marvel | +1939 | +Mystique | +bad | +female | +
+ +### 左连接和右连接(出版商 × 超级英雄) + +除行和变量顺序外,`publishers`与`superheroes`的左连接等价于`superheroes`与`publishers`的右连接。同样,`publishers`与`superheroes`的右连接等价于`superheroes`与`publishers`的左连接。 + +
+ +### 全连接(出版商 × 超级英雄) + +如你所料,除行和变量顺序外,`publishers`与`superheroes`的全连接等价于`superheroes`与`publishers`的全连接。 + +
+ +### 半连接(出版商 × 超级英雄) + +
| publisher | +yr_founded | +
|---|---|
| DC | +1934 | +
| Marvel | +1939 | +
+ +### 反连接(出版商 × 超级英雄) + +
| publisher | +yr_founded | +
|---|---|
| Image | +1992 | +
+ +### 合并数据集的其他工具(复仇者 × 超级英雄) + +当两个数据集具有相同的列(或行)时,还有其他方法可将它们合并为新数据集。我们已使用过`superheroes`数据集,现在尝试将其与`avengers`数据合并。这两个数据集的行数和列数相同,且列名相同。 + +在下方 “数据> 合并” 标签页的截图中,我们可以看到这两个数据集。此处无需选择变量来合并数据集,`选择变量` 中的任何变量在下方命令中都会被忽略。同样,你可以在 `合并后的数据集` 文本输入框中指定合并后数据集的名称。 + +

+ +### 行绑定 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Thor | +good | +male | +Marvel | +
| Iron Man | +good | +male | +Marvel | +
| Hulk | +good | +male | +Marvel | +
| Hawkeye | +good | +male | +Marvel | +
| Black Widow | +good | +female | +Marvel | +
| Captain America | +good | +male | +Marvel | +
| Magneto | +bad | +male | +Marvel | +
| Magneto | +bad | +male | +Marvel | +
| Storm | +good | +female | +Marvel | +
| Mystique | +bad | +female | +Marvel | +
| Batman | +good | +male | +DC | +
| Joker | +bad | +male | +DC | +
| Catwoman | +bad | +female | +DC | +
| Hellboy | +good | +male | +Dark Horse Comics | +
+ +### 列绑定 + +
| name...1 | +alignment...2 | +gender...3 | +publisher...4 | +name...5 | +alignment...6 | +gender...7 | +publisher...8 | +
|---|---|---|---|---|---|---|---|
| Thor | +good | +male | +Marvel | +Magneto | +bad | +male | +Marvel | +
| Iron Man | +good | +male | +Marvel | +Storm | +good | +female | +Marvel | +
| Hulk | +good | +male | +Marvel | +Mystique | +bad | +female | +Marvel | +
| Hawkeye | +good | +male | +Marvel | +Batman | +good | +male | +DC | +
| Black Widow | +good | +female | +Marvel | +Joker | +bad | +male | +DC | +
| Captain America | +good | +male | +Marvel | +Catwoman | +bad | +female | +DC | +
| Magneto | +bad | +male | +Marvel | +Hellboy | +good | +male | +Dark Horse Comics | +
+ +### 交集 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +
+ +### 并集 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Thor | +good | +male | +Marvel | +
| Iron Man | +good | +male | +Marvel | +
| Hulk | +good | +male | +Marvel | +
| Hawkeye | +good | +male | +Marvel | +
| Black Widow | +good | +female | +Marvel | +
| Captain America | +good | +male | +Marvel | +
| Magneto | +bad | +male | +Marvel | +
| Storm | +good | +female | +Marvel | +
| Mystique | +bad | +female | +Marvel | +
| Batman | +good | +male | +DC | +
| Joker | +bad | +male | +DC | +
| Catwoman | +bad | +female | +DC | +
| Hellboy | +good | +male | +Dark Horse Comics | +
+ +### 差集 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Thor | +good | +male | +Marvel | +
| Iron Man | +good | +male | +Marvel | +
| Hulk | +good | +male | +Marvel | +
| Hawkeye | +good | +male | +Marvel | +
| Black Widow | +good | +female | +Marvel | +
| Captain America | +good | +male | +Marvel | +
+ +### 报告 > Rmd + +通过点击屏幕左下角的图标或按键盘上的`ALT-enter`,向*报告 > Rmd*添加代码以(重新)创建合并后的数据集。 + +更多相关讨论请参见《R for data science》中关于关系数据的章节点击查看和Tidy Explain。 + +### R 函数 + +有关`combine_data`函数的帮助,请参见*数据 > 合并*。 diff --git a/radiant.data/inst/app/tools/help/combine.md b/radiant.data/inst/app/tools/help/combine.md new file mode 100644 index 0000000000000000000000000000000000000000..d42dfc749d7fdc05a12bffabae05f7f503f3e360 --- /dev/null +++ b/radiant.data/inst/app/tools/help/combine.md @@ -0,0 +1,1056 @@ +> 合并两个数据集 + +Radiant 中提供了六种来自 Hadley Wickham 等人开发的dplyr包的 “连接(join)”(或 “合并(merge)”)选项。 + +以下示例改编自 Jenny Bryan 的《dplyr 连接函数速查表》,聚焦三个小型数据集(superheroes、publishers和avengers),以说明 R 和 Radiant 中不同的连接类型及其他数据集合并方式。这些数据也可通过以下链接获取 csv 格式文件: + +* superheroes.csv +* publishers.csv +* avengers.csv + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +
| Storm | +good | +female | +Marvel | +
| Mystique | +bad | +female | +Marvel | +
| Batman | +good | +male | +DC | +
| Joker | +bad | +male | +DC | +
| Catwoman | +bad | +female | +DC | +
| Hellboy | +good | +male | +Dark Horse Comics | +
| publisher | +yr_founded | +
|---|---|
| DC | +1934 | +
| Marvel | +1939 | +
| Image | +1992 | +

+ +### 内连接(超级英雄 × 出版商) + +若 x = 超级英雄数据集,y = 出版商数据集: + +> 内连接返回 x 中与 y 有匹配值的所有行,以及 x 和 y 的所有列。若 x 和 y 之间存在多个匹配,所有匹配组合都会被返回。 + +
| name | +alignment | +gender | +publisher | +yr_founded | +
|---|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +1939 | +
| Storm | +good | +female | +Marvel | +1939 | +
| Mystique | +bad | +female | +Marvel | +1939 | +
| Batman | +good | +male | +DC | +1934 | +
| Joker | +bad | +male | +DC | +1934 | +
| Catwoman | +bad | +female | +DC | +1934 | +

+ +### 左连接(超级英雄 × 出版商) + +> 左连接返回 x 的所有行,以及 x 和 y 的所有列。若 x 和 y 之间存在多个匹配,所有匹配组合都会被返回。 + +
| name | +alignment | +gender | +publisher | +yr_founded | +
|---|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +1939 | +
| Storm | +good | +female | +Marvel | +1939 | +
| Mystique | +bad | +female | +Marvel | +1939 | +
| Batman | +good | +male | +DC | +1934 | +
| Joker | +bad | +male | +DC | +1934 | +
| Catwoman | +bad | +female | +DC | +1934 | +
| Hellboy | +good | +male | +Dark Horse Comics | +NA | +

+ +### 右连接(超级英雄 × 出版商) + +> 右连接返回 y 的所有行,以及 y 和 x 的所有列。若 y 和 x 之间存在多个匹配,所有匹配组合都会被返回。 + +
| name | +alignment | +gender | +publisher | +yr_founded | +
|---|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +1939 | +
| Storm | +good | +female | +Marvel | +1939 | +
| Mystique | +bad | +female | +Marvel | +1939 | +
| Batman | +good | +male | +DC | +1934 | +
| Joker | +bad | +male | +DC | +1934 | +
| Catwoman | +bad | +female | +DC | +1934 | +
| NA | +NA | +NA | +Image | +1992 | +

+ +### 全连接(超级英雄 × 出版商) + +> 全连接合并两个数据集,保留出现在任一数据集中的行和列。 + +
| name | +alignment | +gender | +publisher | +yr_founded | +
|---|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +1939 | +
| Storm | +good | +female | +Marvel | +1939 | +
| Mystique | +bad | +female | +Marvel | +1939 | +
| Batman | +good | +male | +DC | +1934 | +
| Joker | +bad | +male | +DC | +1934 | +
| Catwoman | +bad | +female | +DC | +1934 | +
| Hellboy | +good | +male | +Dark Horse Comics | +NA | +
| NA | +NA | +NA | +Image | +1992 | +

| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +
| Storm | +good | +female | +Marvel | +
| Mystique | +bad | +female | +Marvel | +
| Batman | +good | +male | +DC | +
| Joker | +bad | +male | +DC | +
| Catwoman | +bad | +female | +DC | +
+ +### 反连接(超级英雄 × 出版商) + +> 反连接返回 x 中与 y 无匹配值的所有行,仅保留 x 中的列。 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Hellboy | +good | +male | +Dark Horse Comics | +

+ +### 数据集顺序 + +请注意,所选数据集的顺序可能会影响连接结果。如果我们如下设置 `数据> 合并` 标签页,结果如下: + +

+ +### 内连接(出版商 × 超级英雄) + +
| publisher | +yr_founded | +name | +alignment | +gender | +
|---|---|---|---|---|
| DC | +1934 | +Batman | +good | +male | +
| DC | +1934 | +Joker | +bad | +male | +
| DC | +1934 | +Catwoman | +bad | +female | +
| Marvel | +1939 | +Magneto | +bad | +male | +
| Marvel | +1939 | +Storm | +good | +female | +
| Marvel | +1939 | +Mystique | +bad | +female | +
+ +### 左连接和右连接(出版商 × 超级英雄) + +除行和变量顺序外,`publishers`与`superheroes`的左连接等价于`superheroes`与`publishers`的右连接。同样,`publishers`与`superheroes`的右连接等价于`superheroes`与`publishers`的左连接。 + +
+ +### 全连接(出版商 × 超级英雄) + +如你所料,除行和变量顺序外,`publishers`与`superheroes`的全连接等价于`superheroes`与`publishers`的全连接。 + +
+ +### 半连接(出版商 × 超级英雄) + +
| publisher | +yr_founded | +
|---|---|
| DC | +1934 | +
| Marvel | +1939 | +
+ +### 反连接(出版商 × 超级英雄) + +
| publisher | +yr_founded | +
|---|---|
| Image | +1992 | +
+ +### 合并数据集的其他工具(复仇者 × 超级英雄) + +当两个数据集具有相同的列(或行)时,还有其他方法可将它们合并为新数据集。我们已使用过`superheroes`数据集,现在尝试将其与`avengers`数据合并。这两个数据集的行数和列数相同,且列名相同。 + +在下方 “数据> 合并” 标签页的截图中,我们可以看到这两个数据集。此处无需选择变量来合并数据集,`选择变量` 中的任何变量在下方命令中都会被忽略。同样,你可以在 `合并后的数据集` 文本输入框中指定合并后数据集的名称。 + +

+ +### 行绑定 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Thor | +good | +male | +Marvel | +
| Iron Man | +good | +male | +Marvel | +
| Hulk | +good | +male | +Marvel | +
| Hawkeye | +good | +male | +Marvel | +
| Black Widow | +good | +female | +Marvel | +
| Captain America | +good | +male | +Marvel | +
| Magneto | +bad | +male | +Marvel | +
| Magneto | +bad | +male | +Marvel | +
| Storm | +good | +female | +Marvel | +
| Mystique | +bad | +female | +Marvel | +
| Batman | +good | +male | +DC | +
| Joker | +bad | +male | +DC | +
| Catwoman | +bad | +female | +DC | +
| Hellboy | +good | +male | +Dark Horse Comics | +
+ +### 列绑定 + +
| name...1 | +alignment...2 | +gender...3 | +publisher...4 | +name...5 | +alignment...6 | +gender...7 | +publisher...8 | +
|---|---|---|---|---|---|---|---|
| Thor | +good | +male | +Marvel | +Magneto | +bad | +male | +Marvel | +
| Iron Man | +good | +male | +Marvel | +Storm | +good | +female | +Marvel | +
| Hulk | +good | +male | +Marvel | +Mystique | +bad | +female | +Marvel | +
| Hawkeye | +good | +male | +Marvel | +Batman | +good | +male | +DC | +
| Black Widow | +good | +female | +Marvel | +Joker | +bad | +male | +DC | +
| Captain America | +good | +male | +Marvel | +Catwoman | +bad | +female | +DC | +
| Magneto | +bad | +male | +Marvel | +Hellboy | +good | +male | +Dark Horse Comics | +
+ +### 交集 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Magneto | +bad | +male | +Marvel | +
+ +### 并集 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Thor | +good | +male | +Marvel | +
| Iron Man | +good | +male | +Marvel | +
| Hulk | +good | +male | +Marvel | +
| Hawkeye | +good | +male | +Marvel | +
| Black Widow | +good | +female | +Marvel | +
| Captain America | +good | +male | +Marvel | +
| Magneto | +bad | +male | +Marvel | +
| Storm | +good | +female | +Marvel | +
| Mystique | +bad | +female | +Marvel | +
| Batman | +good | +male | +DC | +
| Joker | +bad | +male | +DC | +
| Catwoman | +bad | +female | +DC | +
| Hellboy | +good | +male | +Dark Horse Comics | +
+ +### 差集 + +
| name | +alignment | +gender | +publisher | +
|---|---|---|---|
| Thor | +good | +male | +Marvel | +
| Iron Man | +good | +male | +Marvel | +
| Hulk | +good | +male | +Marvel | +
| Hawkeye | +good | +male | +Marvel | +
| Black Widow | +good | +female | +Marvel | +
| Captain America | +good | +male | +Marvel | +
+ +### 报告 > Rmd + +通过点击屏幕左下角的图标或按键盘上的`ALT-enter`,向*报告 > Rmd*添加代码以(重新)创建合并后的数据集。 + +更多相关讨论请参见《R for data science》中关于关系数据的章节点击查看和Tidy Explain。 + +### R 函数 + +有关`combine_data`函数的帮助,请参见*数据 > 合并*。 diff --git a/radiant.data/inst/app/tools/help/explore.md b/radiant.data/inst/app/tools/help/explore.md new file mode 100644 index 0000000000000000000000000000000000000000..4334aa7777001bdbf6e9a2351b701092a25bf971 --- /dev/null +++ b/radiant.data/inst/app/tools/help/explore.md @@ -0,0 +1,40 @@ +> 汇总和探索你的数据 + +为数据中的一个或多个变量生成汇总统计量。“数据> 探索” 中最强大的功能是,你可以轻松地按一个或多个其他变量来描述数据。 _数据> 透视_ 标签页最适合生成频数表和汇总单个数值变量,而 “数据 > 探索” 标签页允许你使用各种统计量同时汇总多个变量。 + +例如,如果我们从`diamonds`数据集中选择`price`并点击 `生成表格` 按钮,就可以看到观测数(n)、均值、方差等统计量。此外,通过选择`clarity`作为 `分组变量`,也可以轻松获取每种钻石净度水平的平均价格。 + +> 注意,当分类变量(`factor`)从`数值变量` 下拉菜单中被选中时,若所选函数需要,该变量将被转换为数值变量。如果因子水平是数值型的,这些数值将用于所有计算。由于均值、标准差等统计量对非二元分类变量不适用,这类变量将被转换为 0-1(二元)变量,其中第一个水平编码为 1,其他所有水平编码为 0。 + +生成的汇总表格可以通过点击 “存储” 按钮存储到 Radiant 中。如果你想在 “数据 > 可视化” 中基于汇总数据创建图表,这会很有用。要将表格下载为 csv 格式,点击右上角的下载图标。 + +你可以从 `列标题` 下拉菜单中选择选项,切换不同的列标题。可选择 `应用函数`(如均值、中位数等)、`分组变量`(如价格、克拉等),或(首个)`分组依据` 变量的水平(如 Fair-Ideal)。 + +

++Manage data and state: Load data into Radiant, Save data to disk, Remove a dataset from memory, or Save/Load the state of the app
+
Datasets
+When you first start Radiant a dataset (diamonds) with information on diamond prices is shown.
It is good practice to add a description of the data and variables to each file you use. For the files that are bundled with Radiant you will see a brief overview of the variables etc. below a table of the first 10 rows of the data. To add a description for your own data click the Add/edit data description check-box. A text-input box will open below the table where you can add text in markdown format. The description provided for the diamonds data included with Radiant should serve as a good example. After adding or editing a description click the Update description button.
To rename a dataset loaded in Radiant click the Rename data check box, enter a new name, and click the Rename button
Load data
+The best way to load and save data for use in Radiant (and R) is to use the R-data format (rds or rda). These are binary files that can be stored compactly and read into R quickly. Select rds (or rda) from the Load data of type dropdown and click Browse to locate the file(s) you want to load on your computer.
You can get data from a spreadsheet (e.g., Excel or Google sheets) into Radiant in two ways. First, you can save data from the spreadsheet in csv format and then, in Radiant, choose csv from the Load data of type dropdown. Most likely you will have a header row in the csv file with variable names. If the data are not comma separated you can choose semicolon or tab separated. To load a csv file click ‘Browse’ and locate the file on your computer.
++Note: For Windows users with data that contain multibyte characters please make sure your data are in ANSI format so R(adiant) can load the characters correctly.
+
Alternatively, you can select and copy the data in the spreadsheet using CTRL-C (or CMD-C on mac), go to Radiant, choose clipboard from the Load data of type dropdown, and click the Paste button. This is a short-cut that can be convenient for smaller datasets that are cleanly formatted.
If the data is available in R’s global workspace (e.g., you opened a data set in Rstudio and then started Radiant from the addins menu) you can move (or copy) it to Radiant by selecting from global workspace. Select the data.frame(s) you want to use and click the Load button.
To access all data files bundled with Radiant choose examples from the Load data of type dropdown and then click the Load button. These files are used to illustrate the various data and analysis tools accessible in Radiant. For example, the avengers and publishers data are used to illustrate how to combine data in R(adiant) (i.e., Data > Combine).
If csv data is available online choose csv (url) from the dropdown, paste the url into the text input shown, and press Load. If an rda file is available online choose rda (url) from the dropdown, paste the url into the text input, and press Load.
Save data
+As mentioned above, the most convenient way to get data in and out of Radiant is to use the R-data format (rds or rda). Choose rds (or rda) from the Save data to type dropdown and click the Save button to save the selected dataset to file.
Again, it is good practice to add a description of the data and variables to each file you use. To add a description for your own data click the ‘Add/edit data description’ check-box, add text to the text-input window shown in markdown format, and then click the Update description button. When you save the data as an rds (or rda) file the description you created (or edited) will automatically be added to the file as an attribute.
Getting data from Radiant into a spreadsheet can be achieved in two ways. First, you can save data in csv format and load the file into the spreadsheet (i.e., choose csv from the Save data to type dropdown and click the Save button). Alternatively, you can copy the data from Radiant into the clipboard by choosing clipboard from the dropdown and clicking the Copy button, open the spreadsheet, and paste the data from Radiant using CTRL-V (or CMD-V on mac).
To move or copy data from Radiant into R’s global workspace select to global workspace from the Save data to type dropdown and click the Save button.
Save and load state
+It is convenient to work with state files if you want complete your work at another time, perhaps on another computer, or to review previous work you completed using Radiant. You can save and load the state of the Radiant app just as you would a data file. The state file (extension .rda) will contain (1) the data loaded in Radiant, (2) settings for the analyses you were working on, (3) and any reports or code from the Report menu. To save the current state of the app to your hard-disk click the icon in the navbar and then click Save radiant state file. To load load a previous state click the icon in the navbar and the click Load radiant state file.
You can also share a state file with others that would like to replicate your analyses. As an example, download and then load the state file radiant-state.rda as described above. You will navigate automatically to the Data > Visualize tab and will see a plot. See also the Data > View tab for some additional settings loaded from the state file. There is also a report in Report > Rmd created using the Radiant interface. The html file radiant-state.html contains the output created by clicking the Knit report button.
Loading and saving state also works with Rstudio. If you start Radiant from Rstudio and use and then click Stop, the r_data environment and the r_info and r_state lists will be put into Rstudio’s global workspace. If you start radiant again from the Addins menu it will use r_data, r_info, and r_state to restore state. Also, if you load a state file directly into Rstudio it will be used when you start Radiant.
Use Refresh in the menu in the navbar to return to a clean/new state.
Remove data from memory
+If data are loaded in memory that you no longer need in the current session check the Remove data from memory box. Then select the data to remove and click the Remove data button. One datafile will always remain open.
Using commands to load and save data
+R-code can be used in Report > Rmd or Report > R to load data from a file directly into the active Radiant session. Use register("insert-dataset-name") to add a dataset to the Datasets dropdown. R-code can also be used to extract data from Radiant and save it to disk.
R-functions
+For an overview of related R-functions used by Radiant to load and save data see Data > Manage
+


| Operator | +Description | +Example | +
|---|---|---|
| `<` | +less than | +`price < 5000` | +
| `<=` | +less than or equal to | +`carat <= 2` | +
| `>` | +greater than | +`price > 1000` | +
| `>=` | +greater than or equal to | +`carat >= 2` | +
| `==` | +exactly equal to | +`cut == 'Fair'` | +
| `!=` | +not equal to | +`cut != 'Fair'` | +
| `|` | +x OR y | +`price > 10000 | cut == 'Premium'` | +
| `&` | +x AND y | +`carat < 2 & cut == 'Fair'` | +
| `%in%` | +x is one of y | +`cut %in% c('Fair', 'Good')` | +
| is.na | +is missing | +`is.na(price)` | +
| Operator | +Description | +Example | +
|---|---|---|
| `<` | +less than | +`price < 5000` | +
| `<=` | +less than or equal to | +`carat <= 2` | +
| `>` | +greater than | +`price > 1000` | +
| `>=` | +greater than or equal to | +`carat >= 2` | +
| `==` | +exactly equal to | +`cut == 'Fair'` | +
| `!=` | +not equal to | +`cut != 'Fair'` | +
| `|` | +x OR y | +`price > 10000 | cut == 'Premium'` | +
| `&` | +x AND y | +`carat < 2 & cut == 'Fair'` | +
| `%in%` | +x is one of y | +`cut %in% c('Fair', 'Good')` | +
| is.na | +is missing | +`is.na(price)` | +





